submitted
|
Before 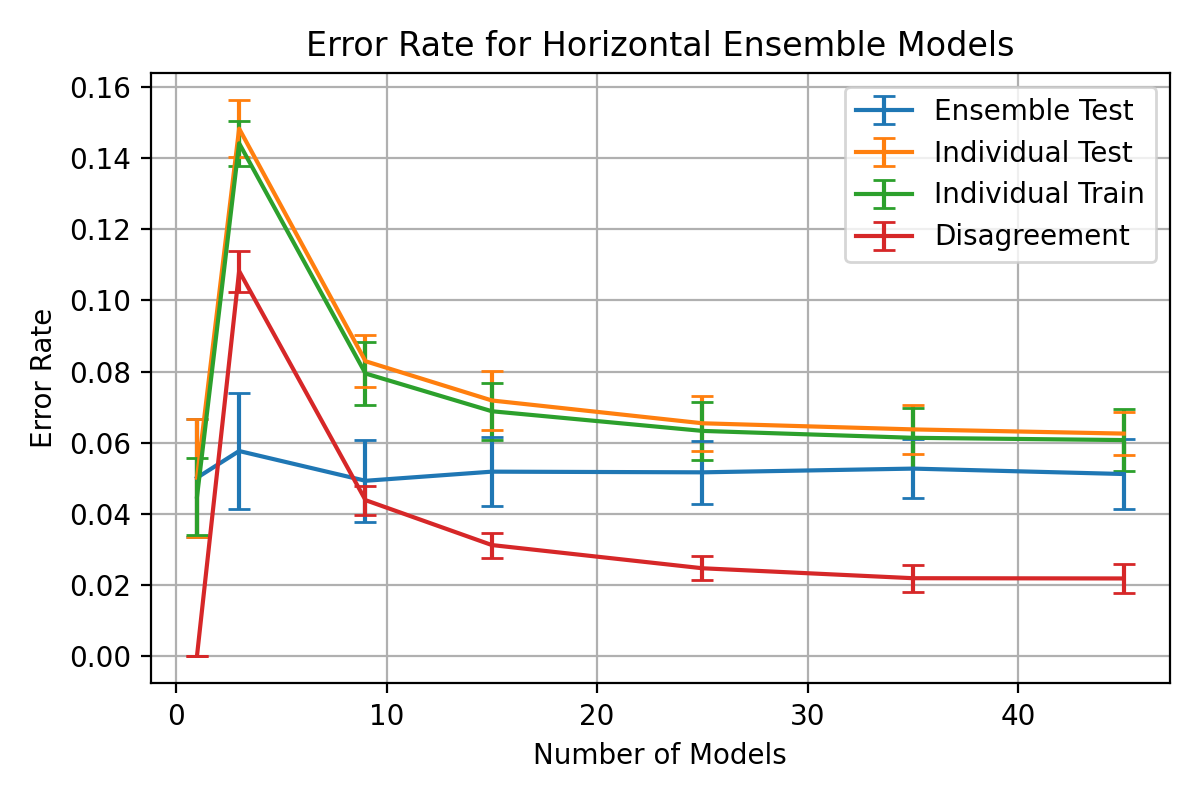
(image error) Size: 94 KiB After 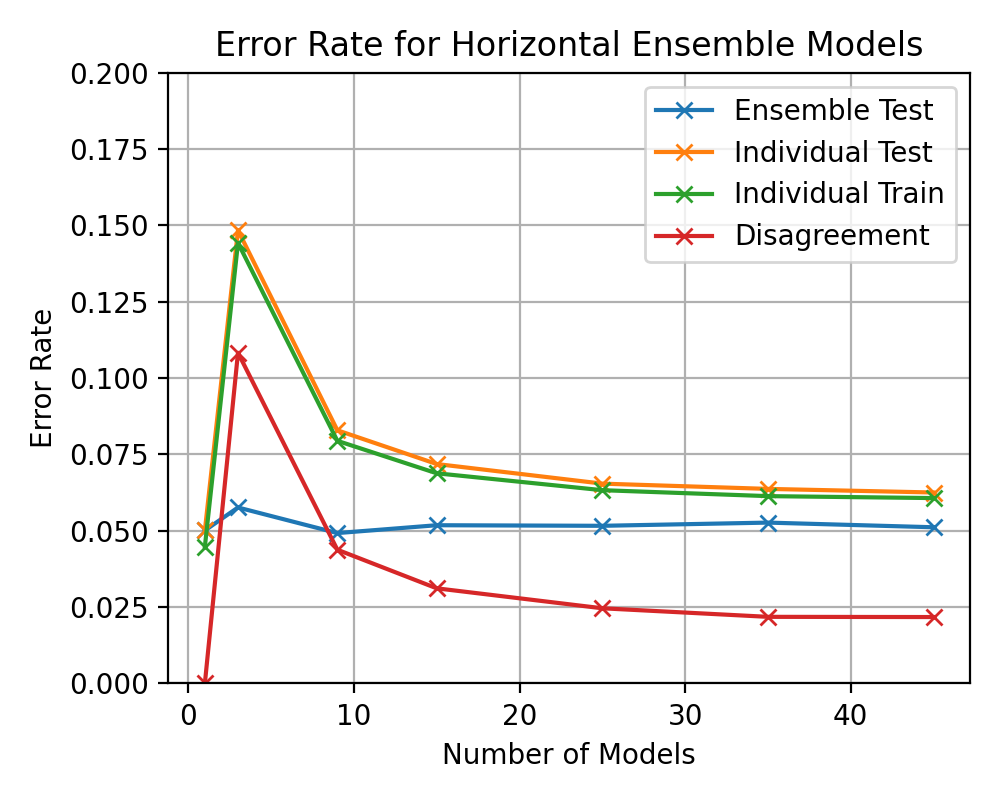
(image error) Size: 86 KiB 

|
BIN
graphs/exp2-test10-error-rate-std.png
Normal file
|
After 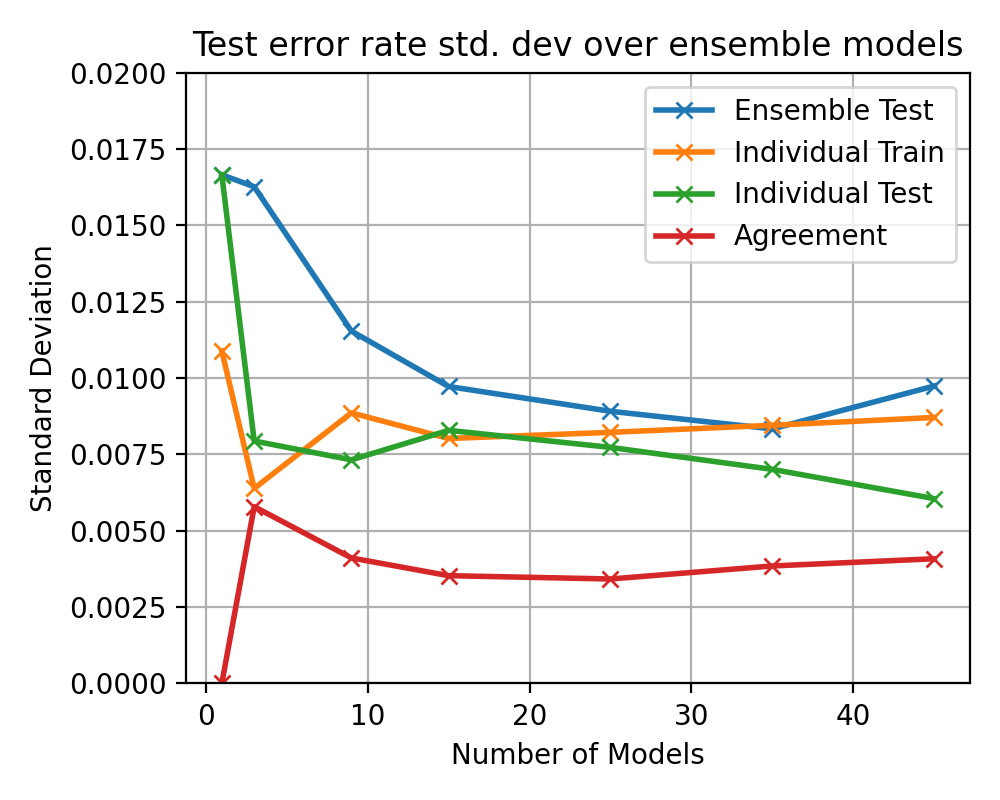
(image error) Size: 92 KiB |
BIN
graphs/exp2-test18-error-rate-curves.png
Normal file
|
After 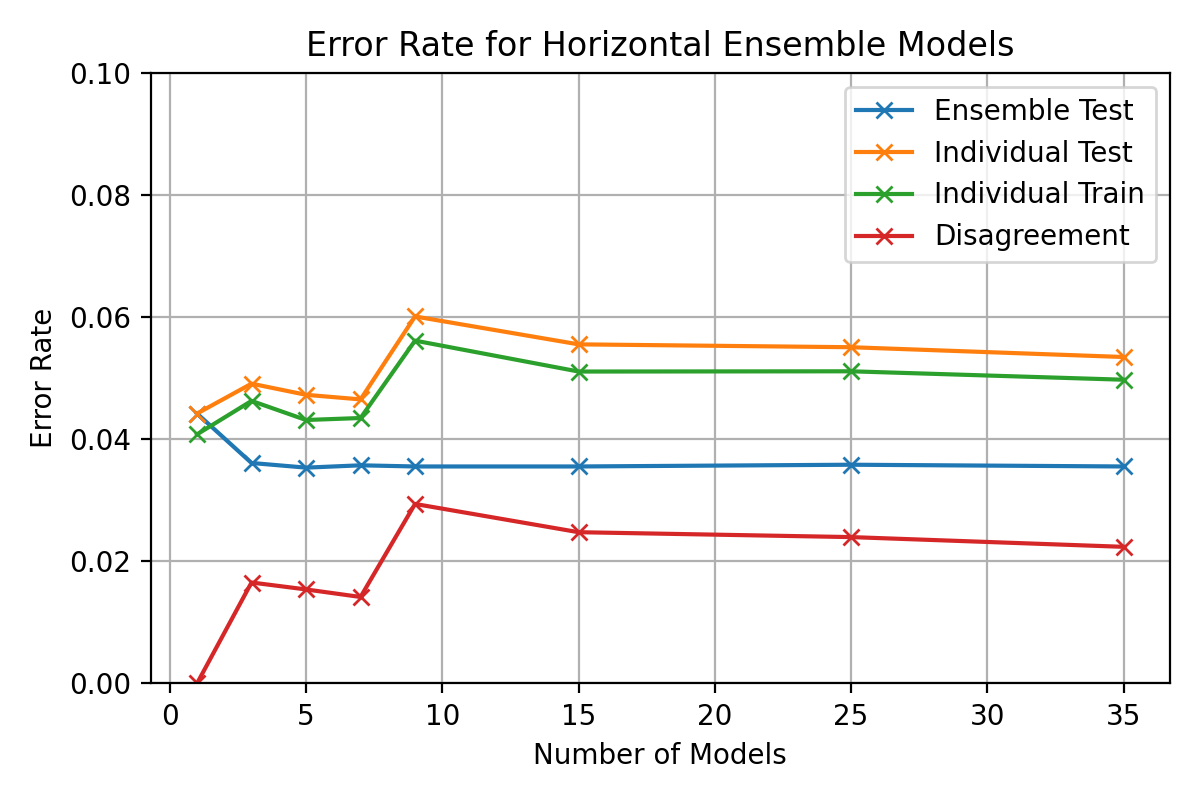
(image error) Size: 77 KiB |
BIN
graphs/exp2-test18-error-rate-std.png
Normal file
|
After 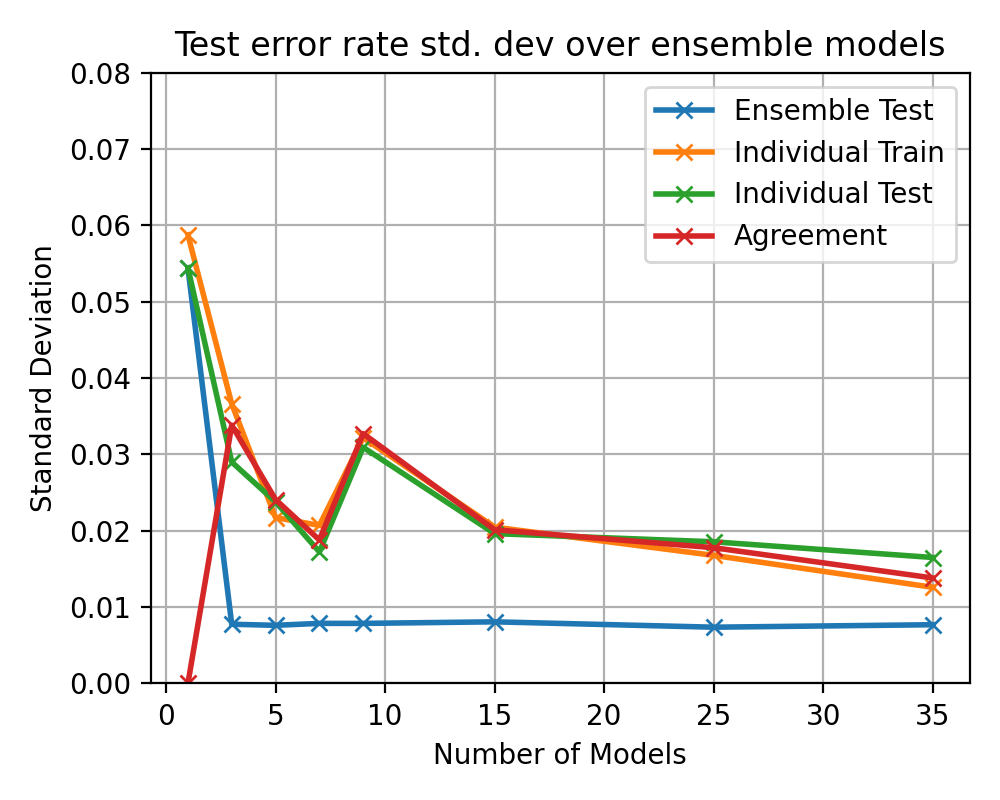
(image error) Size: 90 KiB |
BIN
graphs/exp2-test19-error-rate-curves.png
Normal file
|
After 
(image error) Size: 71 KiB |
BIN
graphs/exp2-test19-error-rate-std.png
Normal file
|
After 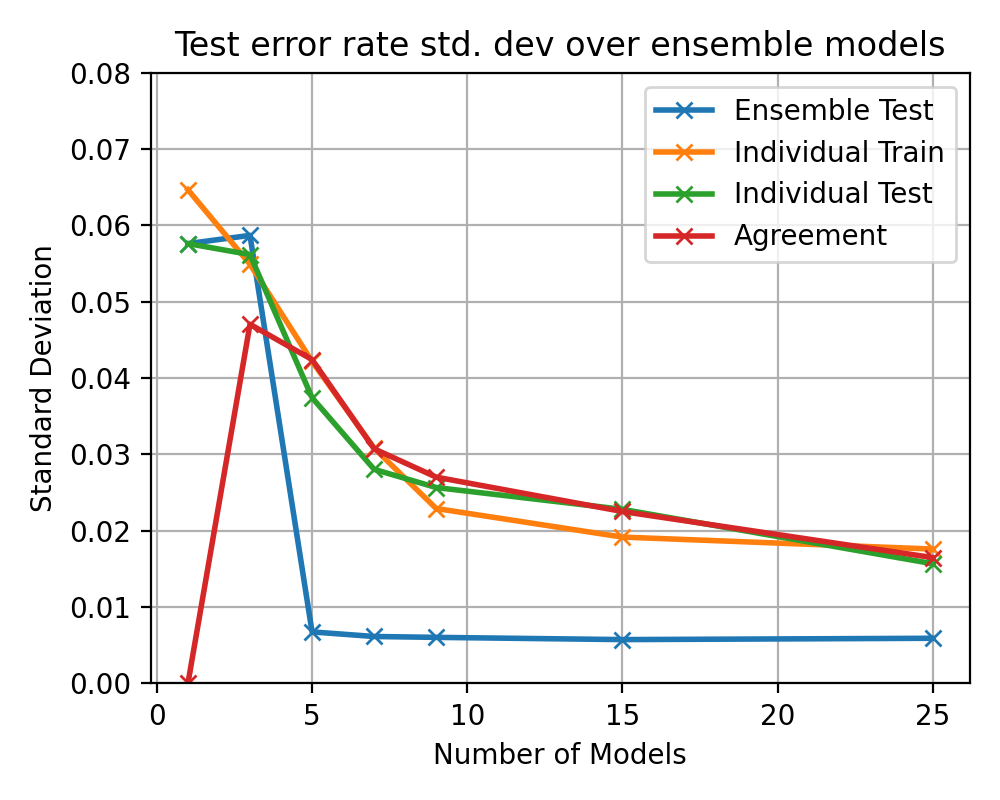
(image error) Size: 89 KiB |
|
Before 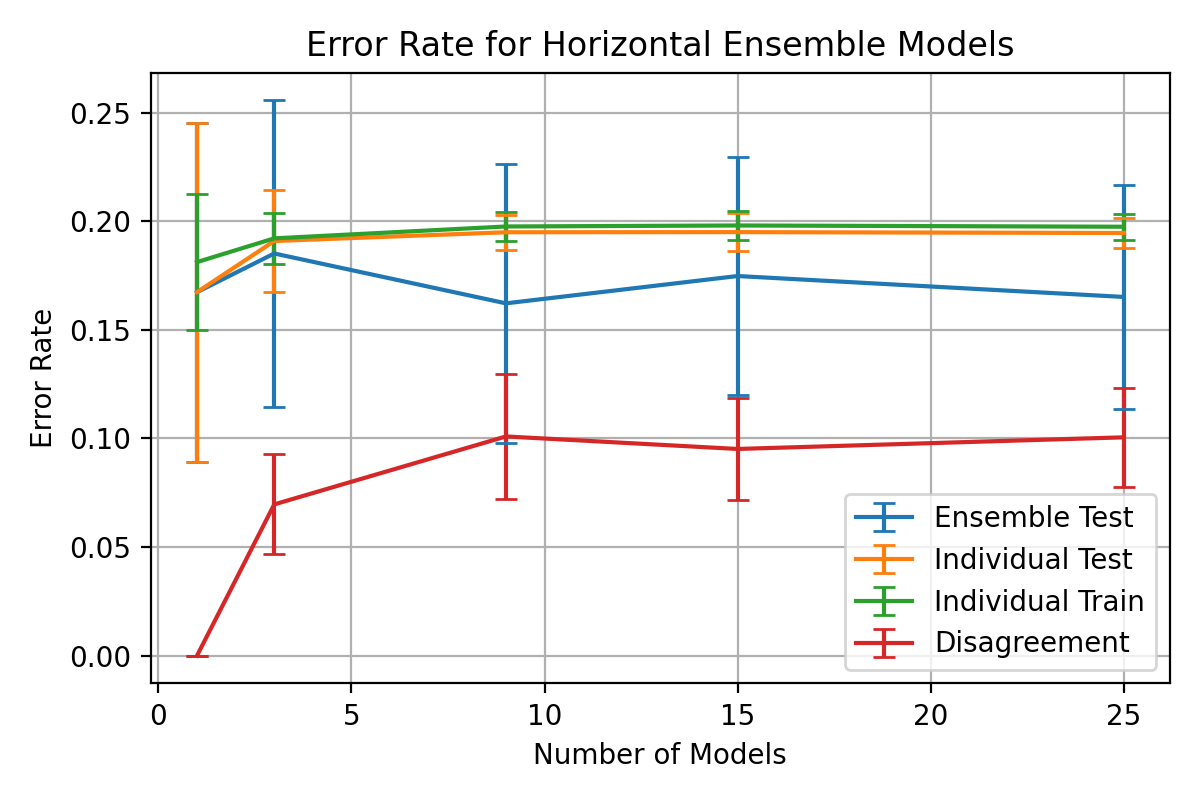
(image error) Size: 68 KiB After 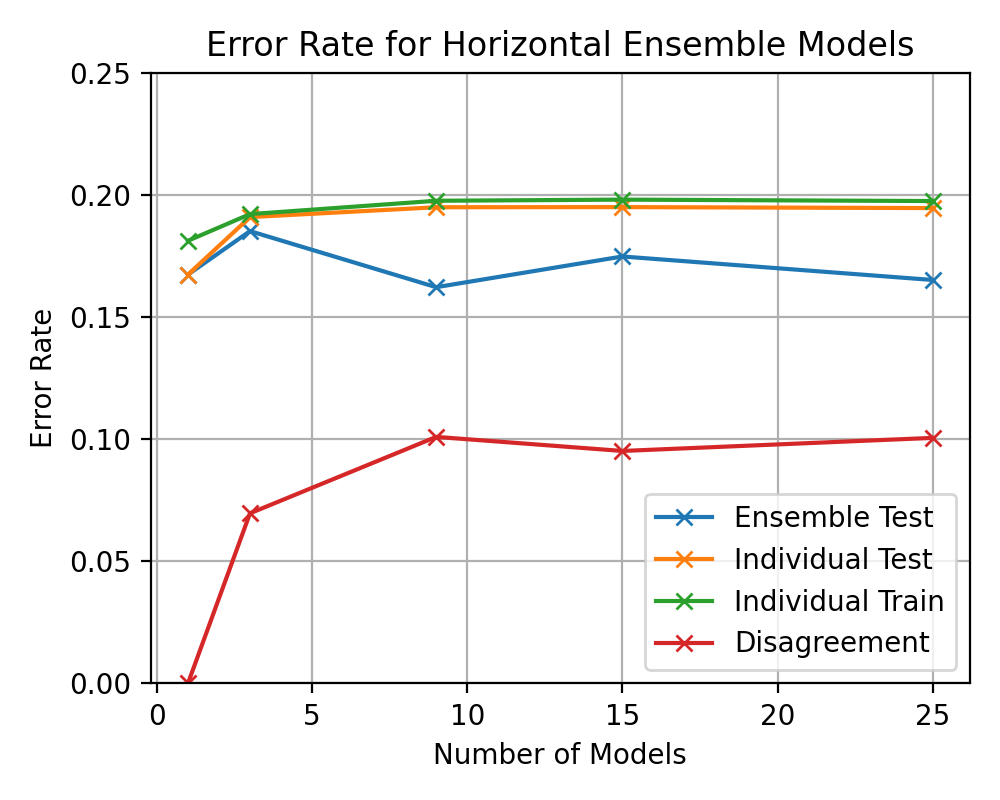
(image error) Size: 68 KiB 

|
BIN
graphs/exp2-test8-error-rate-std.png
Normal file
|
After 
(image error) Size: 80 KiB |
|
Before 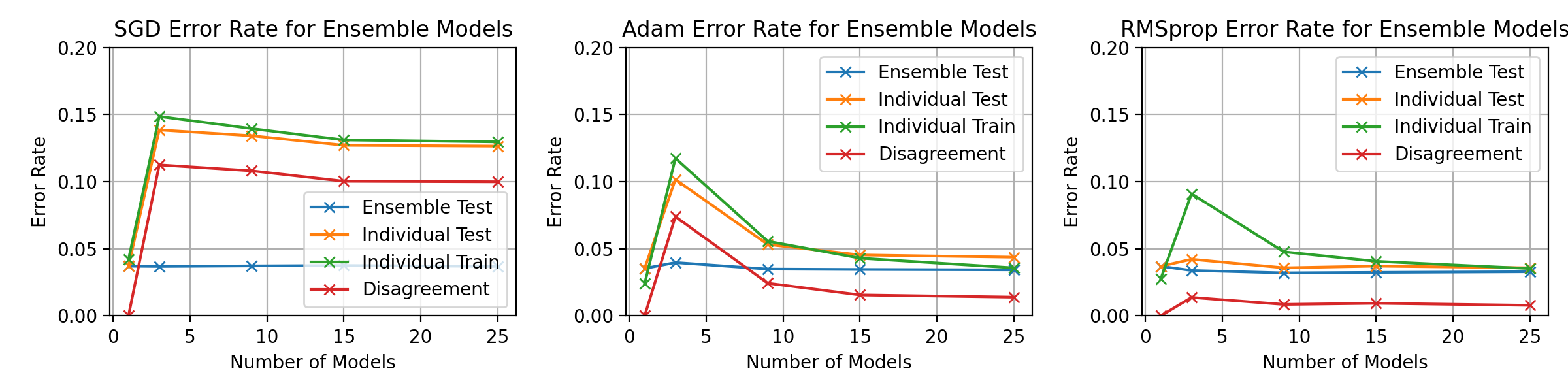
(image error) Size: 140 KiB After 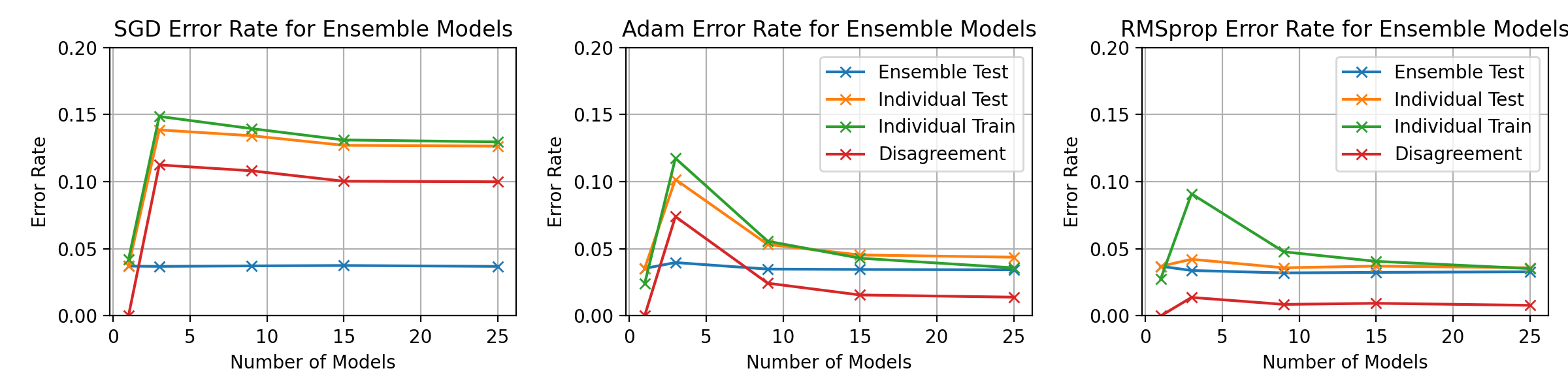
(image error) Size: 122 KiB 

|
BIN
graphs/exp3-test1-errors-rate-std.png
Normal file
|
After 
(image error) Size: 128 KiB |
BIN
graphs/exp3-test10-error-rate-curves.png
Normal file
|
After 
(image error) Size: 120 KiB |
BIN
graphs/exp3-test10-errors-rate-std.png
Normal file
|
After 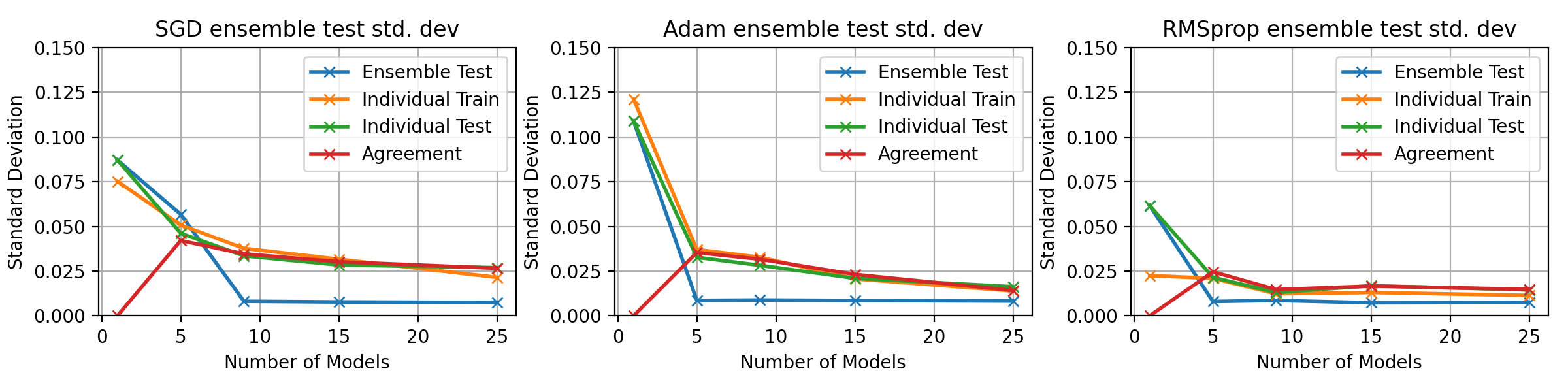
(image error) Size: 130 KiB |
BIN
graphs/exp3-test7-errors-rate-std.png
Normal file
|
After 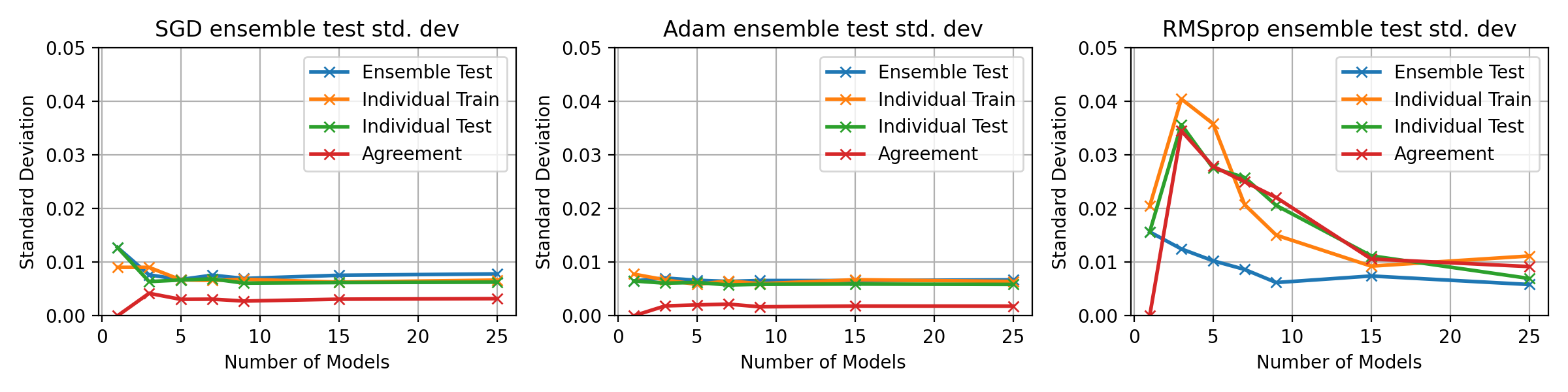
(image error) Size: 120 KiB |
BIN
graphs/exp3-test9-error-rate-curves.png
Normal file
|
After 
(image error) Size: 116 KiB |
BIN
graphs/exp3-test9-errors-rate-std.png
Normal file
|
After 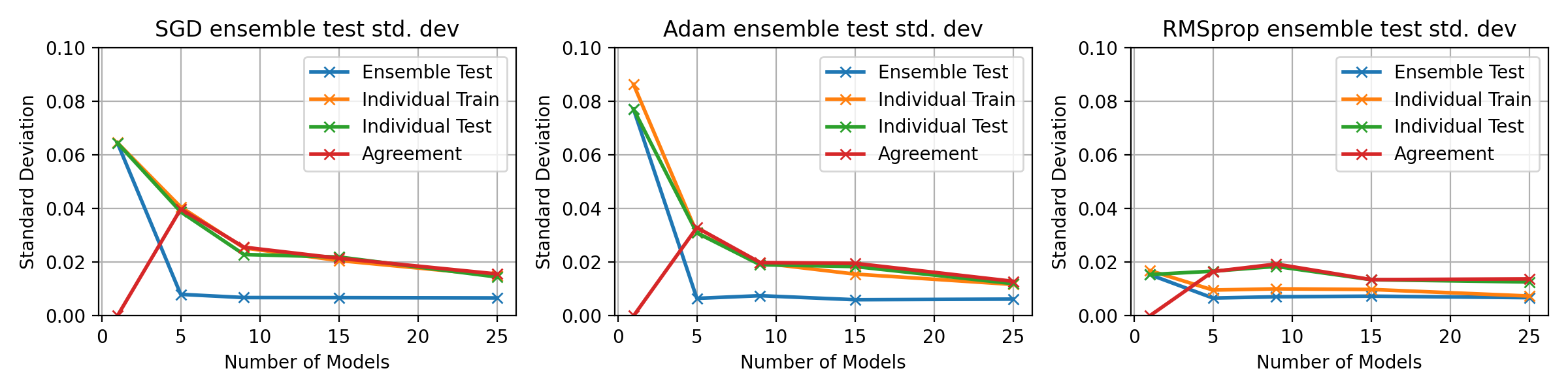
(image error) Size: 126 KiB |
281
nncw.ipynb
294
nncw.py
@ -1,7 +1,7 @@
|
|||||||
#!/usr/bin/env python
|
#!/usr/bin/env python
|
||||||
# coding: utf-8
|
# coding: utf-8
|
||||||
|
|
||||||
# In[2]:
|
# In[41]:
|
||||||
|
|
||||||
|
|
||||||
import numpy as np
|
import numpy as np
|
||||||
@ -19,10 +19,11 @@ import json
|
|||||||
import math
|
import math
|
||||||
import datetime
|
import datetime
|
||||||
import os
|
import os
|
||||||
|
import random
|
||||||
|
|
||||||
from sklearn.model_selection import train_test_split
|
from sklearn.model_selection import train_test_split
|
||||||
|
|
||||||
fig_dpi = 70
|
fig_dpi = 200
|
||||||
|
|
||||||
|
|
||||||
# # Neural Network Training
|
# # Neural Network Training
|
||||||
@ -32,7 +33,7 @@ fig_dpi = 70
|
|||||||
#
|
#
|
||||||
# Read CSVs dumped from MatLab and parse into Pandas DataFrames
|
# Read CSVs dumped from MatLab and parse into Pandas DataFrames
|
||||||
|
|
||||||
# In[3]:
|
# In[42]:
|
||||||
|
|
||||||
|
|
||||||
data = pd.read_csv('features.csv', header=None).T
|
data = pd.read_csv('features.csv', header=None).T
|
||||||
@ -62,7 +63,7 @@ labels.astype(bool).sum(axis=0)
|
|||||||
#
|
#
|
||||||
# Using a 50/50 split
|
# Using a 50/50 split
|
||||||
|
|
||||||
# In[4]:
|
# In[43]:
|
||||||
|
|
||||||
|
|
||||||
data_train, data_test, labels_train, labels_test = train_test_split(data, labels, test_size=0.5
|
data_train, data_test, labels_train, labels_test = train_test_split(data, labels, test_size=0.5
|
||||||
@ -74,7 +75,7 @@ data_train, data_test, labels_train, labels_test = train_test_split(data, labels
|
|||||||
#
|
#
|
||||||
# Get a shallow model with a single hidden layer of varying nodes
|
# Get a shallow model with a single hidden layer of varying nodes
|
||||||
|
|
||||||
# In[5]:
|
# In[44]:
|
||||||
|
|
||||||
|
|
||||||
def get_model(hidden_nodes=9, activation=lambda: 'sigmoid', weight_init=lambda: 'glorot_uniform'):
|
def get_model(hidden_nodes=9, activation=lambda: 'sigmoid', weight_init=lambda: 'glorot_uniform'):
|
||||||
@ -88,7 +89,7 @@ def get_model(hidden_nodes=9, activation=lambda: 'sigmoid', weight_init=lambda:
|
|||||||
|
|
||||||
# Get a Keras Tensorboard callback for dumping data for later analysis
|
# Get a Keras Tensorboard callback for dumping data for later analysis
|
||||||
|
|
||||||
# In[6]:
|
# In[45]:
|
||||||
|
|
||||||
|
|
||||||
def tensorboard_callback(path='tensorboard-logs', prefix=''):
|
def tensorboard_callback(path='tensorboard-logs', prefix=''):
|
||||||
@ -147,7 +148,7 @@ model.metrics[1].result()
|
|||||||
# (Hint2: as epochs increases you can expect the test error rate to reach a minimum and then start increasing, you may need to set the stopping criteria to achieve the desired number of epochs - Hint 3: to find classification error rates for train and test set, you need to check the code from E2, to determine how you may obtain the train and test set patterns)
|
# (Hint2: as epochs increases you can expect the test error rate to reach a minimum and then start increasing, you may need to set the stopping criteria to achieve the desired number of epochs - Hint 3: to find classification error rates for train and test set, you need to check the code from E2, to determine how you may obtain the train and test set patterns)
|
||||||
#
|
#
|
||||||
|
|
||||||
# In[194]:
|
# In[46]:
|
||||||
|
|
||||||
|
|
||||||
# hidden_nodes = [2, 8, 16, 24, 32]
|
# hidden_nodes = [2, 8, 16, 24, 32]
|
||||||
@ -366,6 +367,14 @@ plt.show()
|
|||||||
# |2-4|0.08|0.9|35|1, 2, 8, 16, 32, 64|1, 2, 4, 8, 16, 32, 64, 100| n |
|
# |2-4|0.08|0.9|35|1, 2, 8, 16, 32, 64|1, 2, 4, 8, 16, 32, 64, 100| n |
|
||||||
# |2-5|0.08|0.2|35|1, 2, 8, 16, 32, 64|1, 2, 4, 8, 16, 32, 64, 100| n |
|
# |2-5|0.08|0.2|35|1, 2, 8, 16, 32, 64|1, 2, 4, 8, 16, 32, 64, 100| n |
|
||||||
# |2-6|0.01|0.1|35|2, 8, 16, 24, 32|1, 2, 4, 8, 16, 32, 64, 100, 150, 200| n |
|
# |2-6|0.01|0.1|35|2, 8, 16, 24, 32|1, 2, 4, 8, 16, 32, 64, 100, 150, 200| n |
|
||||||
|
# |2-7|0.01|0.9|35|1, 2, 8, 16, 32, 64|1, 2, 4, 8, 16, 32, 64, 100| n |
|
||||||
|
# |2-8|0.01|0.5|35|1, 2, 8, 16, 32, 64|1, 2, 4, 8, 16, 32, 64, 100| n |
|
||||||
|
# |2-9|0.01|0.3|35|1, 2, 8, 16, 32, 64|1, 2, 4, 8, 16, 32, 64, 100| n |
|
||||||
|
# |2-10|0.01|0.7|35|1, 2, 8, 16, 32, 64|1, 2, 4, 8, 16, 32, 64, 100| n |
|
||||||
|
# |2-11|0.01|0.0|35|1, 2, 8, 16, 32, 64|1, 2, 4, 8, 16, 32, 64, 100| n |
|
||||||
|
# |2-12|0.1|0.0|35|1, 2, 8, 16, 32, 64|1, 2, 4, 8, 16, 32, 64, 100| y |
|
||||||
|
# |2-13|0.5|0.0|35|1, 2, 8, 16, 32, 64|1, 2, 4, 8, 16, 32, 64, 100| y |
|
||||||
|
# |2-14|0.05|0.0|35|1, 2, 8, 16, 32, 64|1, 2, 4, 8, 16, 32, 64, 100| y |
|
||||||
|
|
||||||
# In[214]:
|
# In[214]:
|
||||||
|
|
||||||
@ -396,7 +405,7 @@ for i in range(multi_iterations):
|
|||||||
#
|
#
|
||||||
# (Iterations x [Test/Train] x Number of nodes x Number of epochs)
|
# (Iterations x [Test/Train] x Number of nodes x Number of epochs)
|
||||||
|
|
||||||
# In[301]:
|
# In[268]:
|
||||||
|
|
||||||
|
|
||||||
multi_param_epochs = sorted(list({i["epochs"] for i in multi_param_results[0]}))
|
multi_param_epochs = sorted(list({i["epochs"] for i in multi_param_results[0]}))
|
||||||
@ -420,7 +429,7 @@ print(f'Epochs: {multi_param_epochs}')
|
|||||||
print()
|
print()
|
||||||
print(f'Loss: {multi_param_results[0][0]["loss"]}')
|
print(f'Loss: {multi_param_results[0][0]["loss"]}')
|
||||||
print(f'LR: {multi_param_results[0][0]["optimizer"]["learning_rate"]:.3}')
|
print(f'LR: {multi_param_results[0][0]["optimizer"]["learning_rate"]:.3}')
|
||||||
print(f'Momentum: {multi_param_results[0][0]["optimizer"]["momentum"]:}')
|
print(f'Momentum: {multi_param_results[0][0]["optimizer"]["momentum"]:.3}')
|
||||||
|
|
||||||
|
|
||||||
# #### Export/Import Test Sets
|
# #### Export/Import Test Sets
|
||||||
@ -433,10 +442,10 @@ print(f'Momentum: {multi_param_results[0][0]["optimizer"]["momentum"]:}')
|
|||||||
pickle.dump(multi_param_results, open("results/exp1-test2-3.p", "wb"))
|
pickle.dump(multi_param_results, open("results/exp1-test2-3.p", "wb"))
|
||||||
|
|
||||||
|
|
||||||
# In[300]:
|
# In[267]:
|
||||||
|
|
||||||
|
|
||||||
exp1_testname = 'exp1-test1'
|
exp1_testname = 'exp1-test2-14'
|
||||||
multi_param_results = pickle.load(open(f"results/{exp1_testname}.p", "rb"))
|
multi_param_results = pickle.load(open(f"results/{exp1_testname}.p", "rb"))
|
||||||
|
|
||||||
np.savetxt("exp1-mean.csv", mean_param_accuracy, delimiter=',')
|
np.savetxt("exp1-mean.csv", mean_param_accuracy, delimiter=',')
|
||||||
@ -445,7 +454,7 @@ std_param_accuracy = np.loadtxt("results/test1-exp1-std.csv", delimiter=',')
|
|||||||
# multi_iterations = 30
|
# multi_iterations = 30
|
||||||
# ### Best Results
|
# ### Best Results
|
||||||
|
|
||||||
# In[302]:
|
# In[166]:
|
||||||
|
|
||||||
|
|
||||||
best_param_accuracy_idx = np.unravel_index(np.argmax(mean_param_accuracy[0, :, :]), mean_param_accuracy.shape)
|
best_param_accuracy_idx = np.unravel_index(np.argmax(mean_param_accuracy[0, :, :]), mean_param_accuracy.shape)
|
||||||
@ -453,12 +462,12 @@ best_param_accuracy = mean_param_accuracy[best_param_accuracy_idx]
|
|||||||
best_param_accuracy_nodes = multi_param_nodes[best_param_accuracy_idx[1]]
|
best_param_accuracy_nodes = multi_param_nodes[best_param_accuracy_idx[1]]
|
||||||
best_param_accuracy_epochs = multi_param_epochs[best_param_accuracy_idx[2]]
|
best_param_accuracy_epochs = multi_param_epochs[best_param_accuracy_idx[2]]
|
||||||
|
|
||||||
print(f'Nodes: {best_param_accuracy_nodes}, Epochs: {best_param_accuracy_epochs}, {best_param_accuracy * 100:.1}% Accurate')
|
print(f'Nodes: {best_param_accuracy_nodes}, Epochs: {best_param_accuracy_epochs}, {best_param_accuracy * 100:.3}% Accurate')
|
||||||
|
|
||||||
|
|
||||||
# ### Test Accuracy Surface
|
# ### Test Accuracy Surface
|
||||||
|
|
||||||
# In[303]:
|
# In[269]:
|
||||||
|
|
||||||
|
|
||||||
X, Y = np.meshgrid(multi_param_epochs, multi_param_nodes)
|
X, Y = np.meshgrid(multi_param_epochs, multi_param_nodes)
|
||||||
@ -484,10 +493,10 @@ plt.show()
|
|||||||
|
|
||||||
# ### Test Error Rate Curves
|
# ### Test Error Rate Curves
|
||||||
|
|
||||||
# In[313]:
|
# In[270]:
|
||||||
|
|
||||||
|
|
||||||
fig = plt.figure(figsize=(6, 5))
|
fig = plt.figure(figsize=(5, 4))
|
||||||
# fig = plt.figure()
|
# fig = plt.figure()
|
||||||
fig.set_dpi(fig_dpi)
|
fig.set_dpi(fig_dpi)
|
||||||
|
|
||||||
@ -497,19 +506,42 @@ for idx, (layer, std) in enumerate(zip(mean_param_accuracy[0, :, :], std_param_a
|
|||||||
|
|
||||||
plt.legend()
|
plt.legend()
|
||||||
plt.grid()
|
plt.grid()
|
||||||
plt.title(f"Test error rates for different epochs and hidden nodes")
|
plt.title(f"Test error rates over hidden nodes")
|
||||||
plt.xlabel("Epochs")
|
plt.xlabel("Epochs")
|
||||||
plt.ylabel("Error Rate")
|
plt.ylabel("Error Rate")
|
||||||
plt.ylim(0, 0.6)
|
plt.ylim(0, 0.6)
|
||||||
|
|
||||||
plt.tight_layout()
|
plt.tight_layout()
|
||||||
plt.savefig(f'graphs/{exp1_testname}-error-rate-curves.png')
|
# plt.savefig(f'graphs/{exp1_testname}-error-rate-curves.png')
|
||||||
|
plt.show()
|
||||||
|
|
||||||
|
|
||||||
|
# In[271]:
|
||||||
|
|
||||||
|
|
||||||
|
fig = plt.figure(figsize=(5, 4))
|
||||||
|
# fig = plt.figure()
|
||||||
|
fig.set_dpi(fig_dpi)
|
||||||
|
|
||||||
|
for idx, (layer, std) in enumerate(zip(mean_param_accuracy[0, :, :], std_param_accuracy[0, :, :])):
|
||||||
|
# plt.errorbar(multi_param_epochs, 1 - layer, yerr=std, capsize=4, label=f'{multi_param_nodes[idx]} Nodes')
|
||||||
|
plt.plot(multi_param_epochs, std, 'x-', label=f'{multi_param_nodes[idx]} Nodes', lw=2)
|
||||||
|
|
||||||
|
plt.legend()
|
||||||
|
plt.grid()
|
||||||
|
plt.title(f"Test error rate std. dev over hidden nodes")
|
||||||
|
plt.xlabel("Epochs")
|
||||||
|
plt.ylabel("Standard Deviation")
|
||||||
|
plt.ylim(0, 0.1)
|
||||||
|
|
||||||
|
plt.tight_layout()
|
||||||
|
# plt.savefig(f'graphs/{exp1_testname}-error-rate-std.png')
|
||||||
plt.show()
|
plt.show()
|
||||||
|
|
||||||
|
|
||||||
# ### Test/Train Error Over Nodes
|
# ### Test/Train Error Over Nodes
|
||||||
|
|
||||||
# In[314]:
|
# In[272]:
|
||||||
|
|
||||||
|
|
||||||
fig, axes = plt.subplots(math.ceil(len(multi_param_nodes) / 2), 2, figsize=(6, 6*math.ceil(len(multi_param_nodes) / 2)/3))
|
fig, axes = plt.subplots(math.ceil(len(multi_param_nodes) / 2), 2, figsize=(6, 6*math.ceil(len(multi_param_nodes) / 2)/3))
|
||||||
@ -526,10 +558,10 @@ for idx, (nodes, ax) in enumerate(zip(multi_param_nodes, axes.flatten())):
|
|||||||
ax.grid()
|
ax.grid()
|
||||||
|
|
||||||
fig.tight_layout()
|
fig.tight_layout()
|
||||||
fig.savefig(f'graphs/{exp1_testname}-test-train-error-rate.png')
|
# fig.savefig(f'graphs/{exp1_testname}-test-train-error-rate.png')
|
||||||
|
|
||||||
|
|
||||||
# In[315]:
|
# In[273]:
|
||||||
|
|
||||||
|
|
||||||
fig, axes = plt.subplots(math.ceil(len(multi_param_nodes) / 2), 2, figsize=(6, 6*math.ceil(len(multi_param_nodes) / 2)/3))
|
fig, axes = plt.subplots(math.ceil(len(multi_param_nodes) / 2), 2, figsize=(6, 6*math.ceil(len(multi_param_nodes) / 2)/3))
|
||||||
@ -544,7 +576,7 @@ for idx, (nodes, ax) in enumerate(zip(multi_param_nodes, axes.flatten())):
|
|||||||
ax.grid()
|
ax.grid()
|
||||||
|
|
||||||
fig.tight_layout()
|
fig.tight_layout()
|
||||||
fig.savefig(f'graphs/{exp1_testname}-test-train-error-rate-std.png')
|
# fig.savefig(f'graphs/{exp1_testname}-test-train-error-rate-std.png')
|
||||||
|
|
||||||
|
|
||||||
# # Experiment 2
|
# # Experiment 2
|
||||||
@ -554,7 +586,7 @@ fig.savefig(f'graphs/{exp1_testname}-test-train-error-rate-std.png')
|
|||||||
# (Hint4: to implement majority vote you need to determine the predicted class labels -probably easier to implement yourself rather than use the ensemble matlab functions)
|
# (Hint4: to implement majority vote you need to determine the predicted class labels -probably easier to implement yourself rather than use the ensemble matlab functions)
|
||||||
#
|
#
|
||||||
|
|
||||||
# In[113]:
|
# In[249]:
|
||||||
|
|
||||||
|
|
||||||
num_models=[1, 3, 9, 15, 25]
|
num_models=[1, 3, 9, 15, 25]
|
||||||
@ -562,6 +594,8 @@ num_models=[1, 3, 9, 15, 25]
|
|||||||
def evaluate_ensemble_vote(hidden_nodes=16,
|
def evaluate_ensemble_vote(hidden_nodes=16,
|
||||||
epochs=50,
|
epochs=50,
|
||||||
batch_size=128,
|
batch_size=128,
|
||||||
|
learning_rates=None,
|
||||||
|
rand_ranges=False,
|
||||||
optimizer=lambda: 'sgd',
|
optimizer=lambda: 'sgd',
|
||||||
weight_init=lambda: 'glorot_uniform',
|
weight_init=lambda: 'glorot_uniform',
|
||||||
loss=lambda: 'categorical_crossentropy',
|
loss=lambda: 'categorical_crossentropy',
|
||||||
@ -582,10 +616,12 @@ def evaluate_ensemble_vote(hidden_nodes=16,
|
|||||||
dtest=data_test,
|
dtest=data_test,
|
||||||
ltrain=labels_train,
|
ltrain=labels_train,
|
||||||
ltest=labels_test):
|
ltest=labels_test):
|
||||||
for m in nmodels:
|
|
||||||
|
for m in nmodels: # iterate over different ensemble sizes
|
||||||
if print_params:
|
if print_params:
|
||||||
print(f"Models: {m}")
|
print(f"Models: {m}")
|
||||||
|
|
||||||
|
# response dict object for test stats
|
||||||
response = {"epochs": list(),
|
response = {"epochs": list(),
|
||||||
"num_models": m}
|
"num_models": m}
|
||||||
|
|
||||||
@ -594,29 +630,72 @@ def evaluate_ensemble_vote(hidden_nodes=16,
|
|||||||
###################
|
###################
|
||||||
if isinstance(hidden_nodes, tuple): # for range of hidden nodes, calculate value per model
|
if isinstance(hidden_nodes, tuple): # for range of hidden nodes, calculate value per model
|
||||||
if m == 1:
|
if m == 1:
|
||||||
models = [get_model(int(np.mean(hidden_nodes)), weight_init=weight_init)]
|
if not rand_ranges:
|
||||||
response["nodes"] = [int(np.mean(hidden_nodes))]
|
# just average provided range
|
||||||
|
models = [get_model(int(np.mean(hidden_nodes)), weight_init=weight_init)]
|
||||||
|
response["nodes"] = [int(np.mean(hidden_nodes))]
|
||||||
|
else:
|
||||||
|
# get random val
|
||||||
|
node_val = random.randint(*hidden_nodes)
|
||||||
|
models = [get_model(node_val, weight_init=weight_init)]
|
||||||
|
response["nodes"] = [node_val]
|
||||||
else:
|
else:
|
||||||
models = [get_model(int(i), weight_init=weight_init)
|
if not rand_ranges:
|
||||||
for i in np.linspace(*hidden_nodes, num=m)]
|
# use linspace to generate equally spaced nodes throughout range
|
||||||
response["nodes"] = [int(i) for i in np.linspace(*hidden_nodes, num=m)]
|
models = [get_model(int(i), weight_init=weight_init)
|
||||||
|
for i in np.linspace(*hidden_nodes, num=m)]
|
||||||
|
response["nodes"] = [int(i) for i in np.linspace(*hidden_nodes, num=m)]
|
||||||
|
else:
|
||||||
|
# use random to generate nodes throughout range
|
||||||
|
node_val = [random.randint(*hidden_nodes) for _ in range(m)]
|
||||||
|
models = [get_model(i, weight_init=weight_init) for i in node_val]
|
||||||
|
response["nodes"] = node_val
|
||||||
|
|
||||||
elif hidden_nodes == 'm':
|
elif hidden_nodes == 'm':
|
||||||
|
# incrementing mode, number of nodes ranges from 1 to m
|
||||||
|
# more nodes in larger ensembles
|
||||||
models = [get_model(i+1, weight_init=weight_init) for i in range(m)]
|
models = [get_model(i+1, weight_init=weight_init) for i in range(m)]
|
||||||
response["nodes"] = [i+1 for i in range(m)]
|
response["nodes"] = [i+1 for i in range(m)]
|
||||||
else: # not a range of epochs, just set to given value
|
else:
|
||||||
|
# not a range of epochs, just set to given value
|
||||||
models = [get_model(hidden_nodes, weight_init=weight_init) for _ in range(m)]
|
models = [get_model(hidden_nodes, weight_init=weight_init) for _ in range(m)]
|
||||||
response["nodes"] = hidden_nodes
|
response["nodes"] = hidden_nodes
|
||||||
|
|
||||||
for model in models:
|
######################
|
||||||
model.compile(
|
## COMPILE MODELS
|
||||||
optimizer=optimizer(),
|
######################
|
||||||
loss=loss(),
|
if learning_rates is None:
|
||||||
metrics=metrics
|
# default, just load optimiser
|
||||||
)
|
for model in models:
|
||||||
|
model.compile(
|
||||||
|
optimizer=optimizer(),
|
||||||
|
loss=loss(),
|
||||||
|
metrics=metrics
|
||||||
|
)
|
||||||
|
else:
|
||||||
|
for idx, model in enumerate(models):
|
||||||
|
optim = optimizer()
|
||||||
|
|
||||||
|
# generate learning rate either randomly or linearly
|
||||||
|
if isinstance(learning_rates, tuple):
|
||||||
|
if not rand_ranges:
|
||||||
|
# get equal spaced learning rates
|
||||||
|
optim.learning_rate = np.linspace(*learning_rates, num=m)[idx]
|
||||||
|
else:
|
||||||
|
# get random learning rate
|
||||||
|
optim.learning_rate = random.uniform(*learning_rates)
|
||||||
|
elif learning_rates == '+':
|
||||||
|
# incrementing mode, scale with size of ensemble
|
||||||
|
optim.learning_rate = 0.01 * (idx + 1)
|
||||||
|
|
||||||
|
model.compile(
|
||||||
|
optimizer=optim,
|
||||||
|
loss=loss(),
|
||||||
|
metrics=metrics
|
||||||
|
)
|
||||||
|
|
||||||
if tboard:
|
if tboard:
|
||||||
|
# include a tensorboard callback to dump stats for later analysis
|
||||||
if callbacks is not None:
|
if callbacks is not None:
|
||||||
cb = [i() for i in callbacks] + [tensorboard_callback(prefix=f'exp{exp}-{m}-')]
|
cb = [i() for i in callbacks] + [tensorboard_callback(prefix=f'exp{exp}-{m}-')]
|
||||||
else:
|
else:
|
||||||
@ -627,13 +706,18 @@ def evaluate_ensemble_vote(hidden_nodes=16,
|
|||||||
###################
|
###################
|
||||||
histories = list()
|
histories = list()
|
||||||
for idx, model in enumerate(models):
|
for idx, model in enumerate(models):
|
||||||
if isinstance(epochs, tuple): # for range of epochs, calculate value per model
|
if isinstance(epochs, tuple):
|
||||||
if m == 1:
|
# for range of epochs, calculate value per model
|
||||||
e = np.mean(epochs) # average, not lower bound if single model
|
if not rand_ranges:
|
||||||
|
if m == 1:
|
||||||
|
e = np.mean(epochs) # average, not lower bound if single model
|
||||||
|
else:
|
||||||
|
e = np.linspace(*epochs, num=m)[idx]
|
||||||
|
e = int(e)
|
||||||
else:
|
else:
|
||||||
e = np.linspace(*epochs, num=m)[idx]
|
e = random.randint(*epochs)
|
||||||
e = int(e)
|
else:
|
||||||
else: # not a range of epochs, just set to given value
|
# not a range of epochs, just set to given value
|
||||||
e = epochs
|
e = epochs
|
||||||
|
|
||||||
# print(m, e) # debug
|
# print(m, e) # debug
|
||||||
@ -648,9 +732,9 @@ def evaluate_ensemble_vote(hidden_nodes=16,
|
|||||||
histories.append(history.history)
|
histories.append(history.history)
|
||||||
response["epochs"].append(e)
|
response["epochs"].append(e)
|
||||||
|
|
||||||
########################
|
############################
|
||||||
## FEEDFORWARD TEST
|
## FEEDFORWARD TEST DATA
|
||||||
########################
|
############################
|
||||||
# TEST DATA PREDICTIONS
|
# TEST DATA PREDICTIONS
|
||||||
response["predictions"] = [model(dtest.to_numpy()) for model in models]
|
response["predictions"] = [model(dtest.to_numpy()) for model in models]
|
||||||
# TEST LABEL TENSOR
|
# TEST LABEL TENSOR
|
||||||
@ -665,7 +749,7 @@ def evaluate_ensemble_vote(hidden_nodes=16,
|
|||||||
# take argmax for ensemble predicted class
|
# take argmax for ensemble predicted class
|
||||||
|
|
||||||
correct = 0 # number of correct ensemble predictions
|
correct = 0 # number of correct ensemble predictions
|
||||||
correct_num_models = 0 # when correctly predicted ensembley, proportion of models correctly classifying
|
correct_num_models = 0 # when correctly predicted ensembley, number of models correctly classifying
|
||||||
individual_accuracy = 0 # proportion of models correctly classifying
|
individual_accuracy = 0 # proportion of models correctly classifying
|
||||||
|
|
||||||
# pc = predicted class, pcr = rounded predicted class, gt = ground truth
|
# pc = predicted class, pcr = rounded predicted class, gt = ground truth
|
||||||
@ -712,19 +796,21 @@ def evaluate_ensemble_vote(hidden_nodes=16,
|
|||||||
# ## Single Iteration
|
# ## Single Iteration
|
||||||
# Run a single iteration of ensemble model investigations
|
# Run a single iteration of ensemble model investigations
|
||||||
|
|
||||||
# In[224]:
|
# In[250]:
|
||||||
|
|
||||||
|
|
||||||
single_ensem_results = list()
|
single_ensem_results = list()
|
||||||
# for test in evaluate_ensemble_vote(epochs=(5, 300), optimizer=lambda: tf.keras.optimizers.SGD(learning_rate=0.02)):
|
# for test in evaluate_ensemble_vote(epochs=(5, 300), optimizer=lambda: tf.keras.optimizers.SGD(learning_rate=0.02)):
|
||||||
for test in evaluate_ensemble_vote(hidden_nodes=(1, 400),
|
for test in evaluate_ensemble_vote(hidden_nodes=(1, 20),
|
||||||
epochs=20,
|
epochs=(1, 20),
|
||||||
|
rand_ranges=True,
|
||||||
|
learning_rates=(0.01, 0.5),
|
||||||
optimizer=lambda: tf.keras.optimizers.SGD(learning_rate=0.02)):
|
optimizer=lambda: tf.keras.optimizers.SGD(learning_rate=0.02)):
|
||||||
single_ensem_results.append(test)
|
single_ensem_results.append(test)
|
||||||
print(test["nodes"], test["epochs"])
|
print(test["nodes"], test["epochs"])
|
||||||
|
|
||||||
|
|
||||||
# In[225]:
|
# In[251]:
|
||||||
|
|
||||||
|
|
||||||
fig = plt.figure(figsize=(8, 5))
|
fig = plt.figure(figsize=(8, 5))
|
||||||
@ -774,6 +860,8 @@ plt.show()
|
|||||||
# |15|0.01|0.9|35|50 - 100|50|1, 3, 5, 7, 9, 15, 25, 35, 45| n |
|
# |15|0.01|0.9|35|50 - 100|50|1, 3, 5, 7, 9, 15, 25, 35, 45| n |
|
||||||
# |16|0.01|0.1|35|50 - 100|50|1, 3, 5, 7, 9, 15, 25, 35, 45| n |
|
# |16|0.01|0.1|35|50 - 100|50|1, 3, 5, 7, 9, 15, 25, 35, 45| n |
|
||||||
# |17|0.1|0.1|35|50 - 100|50 - 100|1, 3, 5, 7, 9, 15, 25, 35, 45| n |
|
# |17|0.1|0.1|35|50 - 100|50 - 100|1, 3, 5, 7, 9, 15, 25, 35, 45| n |
|
||||||
|
# |18 (r)|0.01 - 1|0.0|35|1 - 50|20 - 70|1, 3, 5, 7, 9, 15, 25, 35| n |
|
||||||
|
# |19 (r)|0.01 - 1|0.0|35|1 - 100|10 - 70|1, 3, 5, 7, 9, 15, 25| n |
|
||||||
|
|
||||||
# In[335]:
|
# In[335]:
|
||||||
|
|
||||||
@ -829,7 +917,7 @@ for i in range(multi_ensem_iterations):
|
|||||||
# 2. Individual Accuracy
|
# 2. Individual Accuracy
|
||||||
# 3. Agreement
|
# 3. Agreement
|
||||||
|
|
||||||
# In[322]:
|
# In[253]:
|
||||||
|
|
||||||
|
|
||||||
def test_tensor_data(test):
|
def test_tensor_data(test):
|
||||||
@ -839,7 +927,7 @@ def test_tensor_data(test):
|
|||||||
test["agreement"]]
|
test["agreement"]]
|
||||||
|
|
||||||
|
|
||||||
# In[362]:
|
# In[354]:
|
||||||
|
|
||||||
|
|
||||||
multi_ensem_models = sorted(list({i["num_models"] for i in multi_ensem_results[0]}))
|
multi_ensem_models = sorted(list({i["num_models"] for i in multi_ensem_results[0]}))
|
||||||
@ -874,10 +962,10 @@ exp2_testname = 'exp2-test17'
|
|||||||
pickle.dump(multi_ensem_results, open(f"results/{exp2_testname}.p", "wb"))
|
pickle.dump(multi_ensem_results, open(f"results/{exp2_testname}.p", "wb"))
|
||||||
|
|
||||||
|
|
||||||
# In[349]:
|
# In[353]:
|
||||||
|
|
||||||
|
|
||||||
exp2_testname = 'exp2-test16'
|
exp2_testname = 'exp2-test19'
|
||||||
multi_ensem_results = pickle.load(open(f"results/{exp2_testname}.p", "rb"))
|
multi_ensem_results = pickle.load(open(f"results/{exp2_testname}.p", "rb"))
|
||||||
|
|
||||||
np.savetxt("exp2-mean.csv", mean_ensem_accuracy, delimiter=',')
|
np.savetxt("exp2-mean.csv", mean_ensem_accuracy, delimiter=',')
|
||||||
@ -885,7 +973,7 @@ np.savetxt("exp2-std.csv", std_ensem_accuracy, delimiter=',')mean_ensem_accuracy
|
|||||||
std_ensem_accuracy = np.loadtxt("results/test1-exp2-std.csv", delimiter=',')
|
std_ensem_accuracy = np.loadtxt("results/test1-exp2-std.csv", delimiter=',')
|
||||||
# ### Best Results
|
# ### Best Results
|
||||||
|
|
||||||
# In[363]:
|
# In[355]:
|
||||||
|
|
||||||
|
|
||||||
best_ensem_accuracy_idx = np.unravel_index(np.argmax(mean_ensem_accuracy[0, :]), mean_ensem_accuracy.shape)
|
best_ensem_accuracy_idx = np.unravel_index(np.argmax(mean_ensem_accuracy[0, :]), mean_ensem_accuracy.shape)
|
||||||
@ -897,24 +985,24 @@ print(f'Models: {best_ensem_accuracy_models}, {best_ensem_accuracy * 100:.3}% Ac
|
|||||||
|
|
||||||
# ### Test/Train Error Over Model Numbers
|
# ### Test/Train Error Over Model Numbers
|
||||||
|
|
||||||
# In[364]:
|
# In[356]:
|
||||||
|
|
||||||
|
|
||||||
fig = plt.figure(figsize=(6, 4))
|
fig = plt.figure(figsize=(5, 4))
|
||||||
fig.set_dpi(fig_dpi)
|
fig.set_dpi(fig_dpi)
|
||||||
|
|
||||||
# plt.plot(multi_ensem_models, 1 - mean_ensem_accuracy[0, :], 'x-', label='Ensemble Test')
|
plt.plot(multi_ensem_models, 1 - mean_ensem_accuracy[0, :], 'x-', label='Ensemble Test')
|
||||||
# plt.plot(multi_ensem_models, 1 - mean_ensem_accuracy[2, :], 'x-', label='Individual Test')
|
plt.plot(multi_ensem_models, 1 - mean_ensem_accuracy[2, :], 'x-', label='Individual Test')
|
||||||
# plt.plot(multi_ensem_models, 1 - mean_ensem_accuracy[1, :], 'x-', label='Individual Train')
|
plt.plot(multi_ensem_models, 1 - mean_ensem_accuracy[1, :], 'x-', label='Individual Train')
|
||||||
# plt.plot(multi_ensem_models, 1 - mean_ensem_accuracy[3, :], 'x-', label='Agreement')
|
plt.plot(multi_ensem_models, 1 - mean_ensem_accuracy[3, :], 'x-', label='Disagreement')
|
||||||
|
|
||||||
plt.errorbar(multi_ensem_models, 1 - mean_ensem_accuracy[0, :], yerr=std_ensem_accuracy[0, :], capsize=4, label='Ensemble Test')
|
# plt.errorbar(multi_ensem_models, 1 - mean_ensem_accuracy[0, :], yerr=std_ensem_accuracy[0, :], capsize=4, label='Ensemble Test')
|
||||||
plt.errorbar(multi_ensem_models, 1 - mean_ensem_accuracy[2, :], yerr=std_ensem_accuracy[2, :], capsize=4, label='Individual Test')
|
# plt.errorbar(multi_ensem_models, 1 - mean_ensem_accuracy[2, :], yerr=std_ensem_accuracy[2, :], capsize=4, label='Individual Test')
|
||||||
plt.errorbar(multi_ensem_models, 1 - mean_ensem_accuracy[1, :], yerr=std_ensem_accuracy[1, :], capsize=4, label='Individual Train')
|
# plt.errorbar(multi_ensem_models, 1 - mean_ensem_accuracy[1, :], yerr=std_ensem_accuracy[1, :], capsize=4, label='Individual Train')
|
||||||
plt.errorbar(multi_ensem_models, 1 - mean_ensem_accuracy[3, :], yerr=std_ensem_accuracy[3, :], capsize=4, label='Disagreement')
|
# plt.errorbar(multi_ensem_models, 1 - mean_ensem_accuracy[3, :], yerr=std_ensem_accuracy[3, :], capsize=4, label='Disagreement')
|
||||||
|
|
||||||
plt.title(f"Error Rate for Horizontal Ensemble Models")
|
plt.title(f"Error Rate for Horizontal Ensemble Models")
|
||||||
# plt.ylim(0, 0.2)
|
plt.ylim(0, 0.1)
|
||||||
# plt.ylim(0, np.max(1 - mean_ensem_accuracy + std_ensem_accuracy) + 0.05)
|
# plt.ylim(0, np.max(1 - mean_ensem_accuracy + std_ensem_accuracy) + 0.05)
|
||||||
plt.grid()
|
plt.grid()
|
||||||
plt.legend()
|
plt.legend()
|
||||||
@ -922,11 +1010,35 @@ plt.xlabel("Number of Models")
|
|||||||
plt.ylabel("Error Rate")
|
plt.ylabel("Error Rate")
|
||||||
|
|
||||||
plt.tight_layout()
|
plt.tight_layout()
|
||||||
plt.savefig(f'graphs/{exp2_testname}-error-rate-curves.png')
|
# plt.savefig(f'graphs/{exp2_testname}-error-rate-curves.png')
|
||||||
|
|
||||||
plt.show()
|
plt.show()
|
||||||
|
|
||||||
|
|
||||||
|
# In[305]:
|
||||||
|
|
||||||
|
|
||||||
|
fig = plt.figure(figsize=(5, 4))
|
||||||
|
# fig = plt.figure()
|
||||||
|
fig.set_dpi(fig_dpi)
|
||||||
|
|
||||||
|
plt.plot(multi_ensem_models, std_ensem_accuracy[0, :], 'x-', label='Ensemble Test', lw=2)
|
||||||
|
plt.plot(multi_ensem_models, std_ensem_accuracy[1, :], 'x-', label='Individual Train', lw=2)
|
||||||
|
plt.plot(multi_ensem_models, std_ensem_accuracy[2, :], 'x-', label='Individual Test', lw=2)
|
||||||
|
plt.plot(multi_ensem_models, std_ensem_accuracy[3, :], 'x-', label='Agreement', lw=2)
|
||||||
|
|
||||||
|
plt.legend()
|
||||||
|
plt.grid()
|
||||||
|
plt.title(f"Test error rate std. dev over ensemble models")
|
||||||
|
plt.xlabel("Number of Models")
|
||||||
|
plt.ylabel("Standard Deviation")
|
||||||
|
plt.ylim(0, 0.08)
|
||||||
|
|
||||||
|
plt.tight_layout()
|
||||||
|
# plt.savefig(f'graphs/{exp2_testname}-error-rate-std.png')
|
||||||
|
plt.show()
|
||||||
|
|
||||||
|
|
||||||
# # Experiment 3
|
# # Experiment 3
|
||||||
#
|
#
|
||||||
# Repeat Exp 2) for cancer dataset with two different optimisers of your choice e.g. 'trainlm' and 'trainrp'. Comment and discuss the result and decide which is more appropriate training algorithm for the problem. In your discussion, include in your description a detailed account of how the training algorithms (optimisations) work.
|
# Repeat Exp 2) for cancer dataset with two different optimisers of your choice e.g. 'trainlm' and 'trainrp'. Comment and discuss the result and decide which is more appropriate training algorithm for the problem. In your discussion, include in your description a detailed account of how the training algorithms (optimisations) work.
|
||||||
@ -978,6 +1090,8 @@ for test in evaluate_optimisers(epochs=(5, 300), nmodels=[1, 3, 5]):
|
|||||||
# | 6 | SGD | Adam | RMSprop | 0.02 | 0.01 | 1e7 | 35 | m | 50 | 1, 3, 9, 15, 25, 35, 45 | n |
|
# | 6 | SGD | Adam | RMSprop | 0.02 | 0.01 | 1e7 | 35 | m | 50 | 1, 3, 9, 15, 25, 35, 45 | n |
|
||||||
# | 7 | SGD | Adam | RMSprop | 0.1 | 0.9 | 1e-8 | 35 | 1 - 400 | 50 - 100 | 1, 3, 5, 7, 9, 15, 25 | n |
|
# | 7 | SGD | Adam | RMSprop | 0.1 | 0.9 | 1e-8 | 35 | 1 - 400 | 50 - 100 | 1, 3, 5, 7, 9, 15, 25 | n |
|
||||||
# | 8 | SGD | Adam | RMSprop | 0.05 | 0.9 | 1e-8 | 35 | 1 - 400 | 50 - 100 | 1, 3, 5, 7, 9, 15, 25 | n |
|
# | 8 | SGD | Adam | RMSprop | 0.05 | 0.9 | 1e-8 | 35 | 1 - 400 | 50 - 100 | 1, 3, 5, 7, 9, 15, 25 | n |
|
||||||
|
# | 9 (r) | SGD | Adam | RMSprop | 0.01 - 1 | 0.0 | 1e-7 | 35 | 1 - 100 | 10 - 70 | 1, 5, 9, 15, 25 | n |
|
||||||
|
# | 10 (r) | SGD | Adam | RMSprop | 0.01 - 1 | 0.0 | 1e-7 | 35 | 1 - 100 | 1 - 70 | 1, 5, 9, 15, 25 | n |
|
||||||
|
|
||||||
# In[27]:
|
# In[27]:
|
||||||
|
|
||||||
@ -1027,7 +1141,7 @@ for i in range(multi_optim_iterations):
|
|||||||
# 2. Individual Accuracy
|
# 2. Individual Accuracy
|
||||||
# 3. Agreement
|
# 3. Agreement
|
||||||
|
|
||||||
# In[467]:
|
# In[339]:
|
||||||
|
|
||||||
|
|
||||||
multi_optim_results_dict = dict() # indexed by optimiser name
|
multi_optim_results_dict = dict() # indexed by optimiser name
|
||||||
@ -1085,16 +1199,16 @@ print(f'Loss: {multi_optim_results[0][0][0]["loss"]}')
|
|||||||
pickle.dump(multi_optim_results, open("results/exp3-test5.p", "wb"))
|
pickle.dump(multi_optim_results, open("results/exp3-test5.p", "wb"))
|
||||||
|
|
||||||
|
|
||||||
# In[466]:
|
# In[338]:
|
||||||
|
|
||||||
|
|
||||||
exp3_testname = 'exp3-test8'
|
exp3_testname = 'exp3-test10'
|
||||||
multi_optim_results = pickle.load(open(f"results/{exp3_testname}.p", "rb"))
|
multi_optim_results = pickle.load(open(f"results/{exp3_testname}.p", "rb"))
|
||||||
|
|
||||||
|
|
||||||
# ### Best Results
|
# ### Best Results
|
||||||
|
|
||||||
# In[468]:
|
# In[340]:
|
||||||
|
|
||||||
|
|
||||||
for optim, optim_results in optim_tensors.items():
|
for optim, optim_results in optim_tensors.items():
|
||||||
@ -1107,7 +1221,7 @@ for optim, optim_results in optim_tensors.items():
|
|||||||
|
|
||||||
# ### Optimiser Error Rates
|
# ### Optimiser Error Rates
|
||||||
|
|
||||||
# In[469]:
|
# In[343]:
|
||||||
|
|
||||||
|
|
||||||
fig, axes = plt.subplots(1, 3, figsize=(12, 3))
|
fig, axes = plt.subplots(1, 3, figsize=(12, 3))
|
||||||
@ -1125,11 +1239,11 @@ for idx, ((optimiser_name, tensors_dict), ax) in enumerate(zip(optim_tensors.ite
|
|||||||
# ax.errorbar(multi_optim_models, 1 - tensors_dict["mean"][3, :], yerr=tensors_dict["std"][3, :], capsize=4, label='Disagreement')
|
# ax.errorbar(multi_optim_models, 1 - tensors_dict["mean"][3, :], yerr=tensors_dict["std"][3, :], capsize=4, label='Disagreement')
|
||||||
|
|
||||||
ax.set_title(f"{optimiser_name} Error Rate for Ensemble Models")
|
ax.set_title(f"{optimiser_name} Error Rate for Ensemble Models")
|
||||||
ax.set_ylim(0, 0.1)
|
ax.set_ylim(0, 0.15)
|
||||||
# ax.set_ylim(0, np.max([np.max(1 - i["mean"] + i["std"]) for i in optim_tensors.values()]) + 0.03)
|
# ax.set_ylim(0, np.max([np.max(1 - i["mean"] + i["std"]) for i in optim_tensors.values()]) + 0.03)
|
||||||
ax.grid()
|
ax.grid()
|
||||||
# if idx > 0:
|
# if idx > 0:
|
||||||
ax.legend()
|
# ax.legend()
|
||||||
ax.set_xlabel("Number of Models")
|
ax.set_xlabel("Number of Models")
|
||||||
ax.set_ylabel("Error Rate")
|
ax.set_ylabel("Error Rate")
|
||||||
|
|
||||||
@ -1138,11 +1252,39 @@ axes[1].legend()
|
|||||||
axes[2].legend()
|
axes[2].legend()
|
||||||
|
|
||||||
plt.tight_layout()
|
plt.tight_layout()
|
||||||
plt.savefig(f'graphs/{exp3_testname}-error-rate-curves.png')
|
# plt.savefig(f'graphs/{exp3_testname}-error-rate-curves.png')
|
||||||
|
|
||||||
plt.show()
|
plt.show()
|
||||||
|
|
||||||
|
|
||||||
|
# In[345]:
|
||||||
|
|
||||||
|
|
||||||
|
# fig = plt.figure(figsize=(5, 4))
|
||||||
|
# fig = plt.figure()
|
||||||
|
# fig.set_dpi(fig_dpi)
|
||||||
|
|
||||||
|
fig, axes = plt.subplots(1, 3, figsize=(12, 3))
|
||||||
|
fig.set_dpi(fig_dpi)
|
||||||
|
|
||||||
|
for idx, ((optimiser_name, tensors_dict), ax) in enumerate(zip(optim_tensors.items(), axes.flatten())):
|
||||||
|
ax.plot(multi_optim_models, tensors_dict["std"][0, :], 'x-', label='Ensemble Test', lw=2)
|
||||||
|
ax.plot(multi_optim_models, tensors_dict["std"][1, :], 'x-', label='Individual Train', lw=2)
|
||||||
|
ax.plot(multi_optim_models, tensors_dict["std"][2, :], 'x-', label='Individual Test', lw=2)
|
||||||
|
ax.plot(multi_optim_models, tensors_dict["std"][3, :], 'x-', label='Agreement', lw=2)
|
||||||
|
|
||||||
|
ax.legend()
|
||||||
|
ax.grid()
|
||||||
|
ax.set_title(f"{optimiser_name} ensemble test std. dev")
|
||||||
|
ax.set_xlabel("Number of Models")
|
||||||
|
ax.set_ylabel("Standard Deviation")
|
||||||
|
ax.set_ylim(0, 0.15)
|
||||||
|
|
||||||
|
plt.tight_layout()
|
||||||
|
# plt.savefig(f'graphs/{exp3_testname}-errors-rate-std.png')
|
||||||
|
plt.show()
|
||||||
|
|
||||||
|
|
||||||
# In[ ]:
|
# In[ ]:
|
||||||
|
|
||||||
|
|
||||||
|
|||||||