adding exp1/2 data, writing
24
README.md
@ -5,4 +5,26 @@ Evaluating a neural network using the MatLab `cancer_dataset`. Development conta
|
||||
1. Evaluate the network's tendency to overfit by varying the number of epochs and hidden layers being used
|
||||
2. Multiple classifier performance using majority vote
|
||||
3. Repeat 2 with two different optimisers (`trainlm`, `trainrp`)
|
||||
4. ***Extension***: Distinguish between two equi-probable classes of overlapping 2D Gaussians
|
||||
4. ***Extension***: Distinguish between two equi-probable classes of overlapping 2D Gaussians
|
||||
|
||||

|
||||
|
||||
## Timing
|
||||
|
||||
### exp 1
|
||||
|
||||
CPU: 2min 36s ± 1.66 s per loop (mean ± std. dev. of 2 runs, 2 loops each)
|
||||
|
||||
GPU: 3min 5s ± 2.95 s per loop (mean ± std. dev. of 2 runs, 2 loops each)
|
||||
|
||||
### exp 2
|
||||
|
||||
CPU: 26 s ± 62.9 ms per loop (mean ± std. dev. of 2 runs, 2 loops each)
|
||||
|
||||
GPU: 57.6 s ± 46.7 ms per loop (mean ± std. dev. of 2 runs, 2 loops each)
|
||||
|
||||
### exp 3
|
||||
|
||||
CPU: 1min 19s ± 1.6 s per loop (mean ± std. dev. of 2 runs, 2 loops each)
|
||||
|
||||
GPU: 3min 25s ± 280 ms per loop (mean ± std. dev. of 2 runs, 2 loops each)
|
||||
BIN
graphs/exp1-test1-acc-surf.png
Normal file
|
After 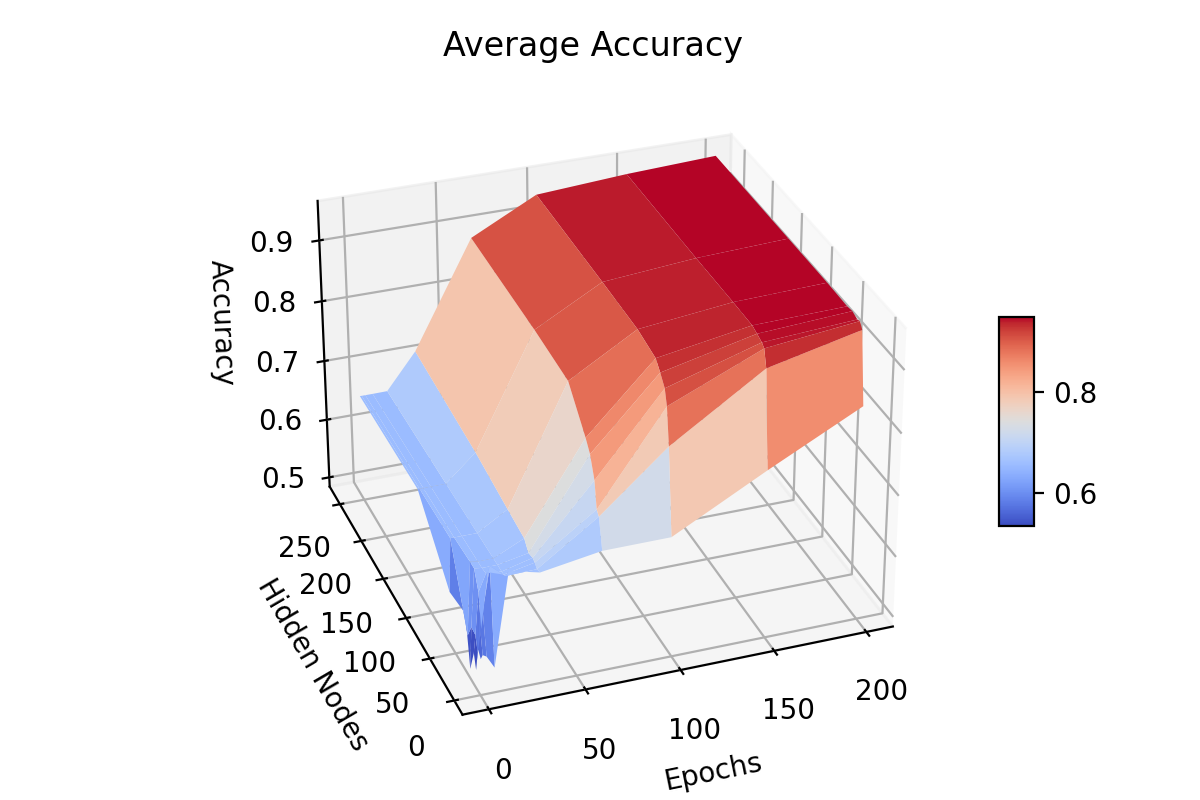
(image error) Size: 154 KiB |
|
Before 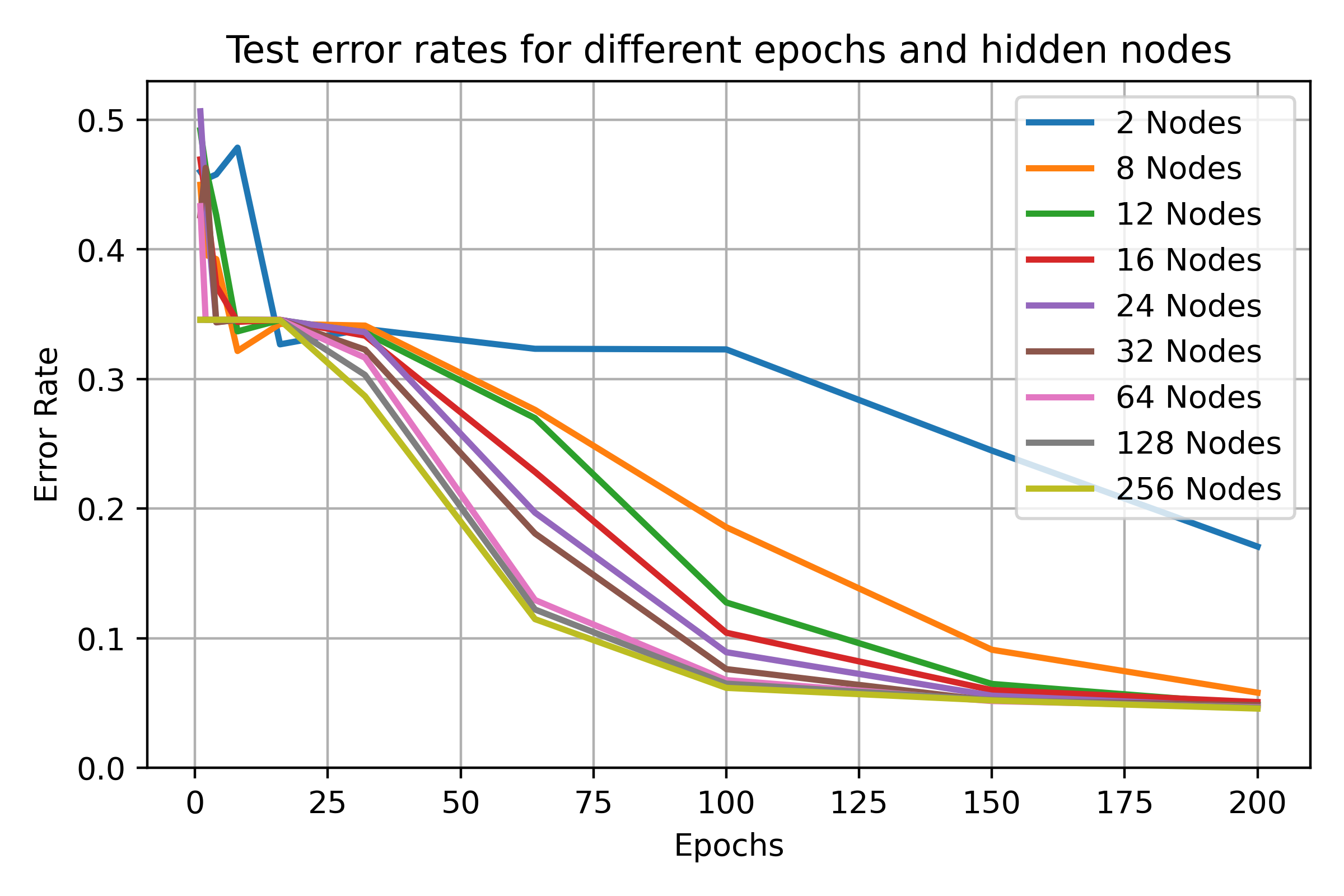
(image error) Size: 335 KiB After 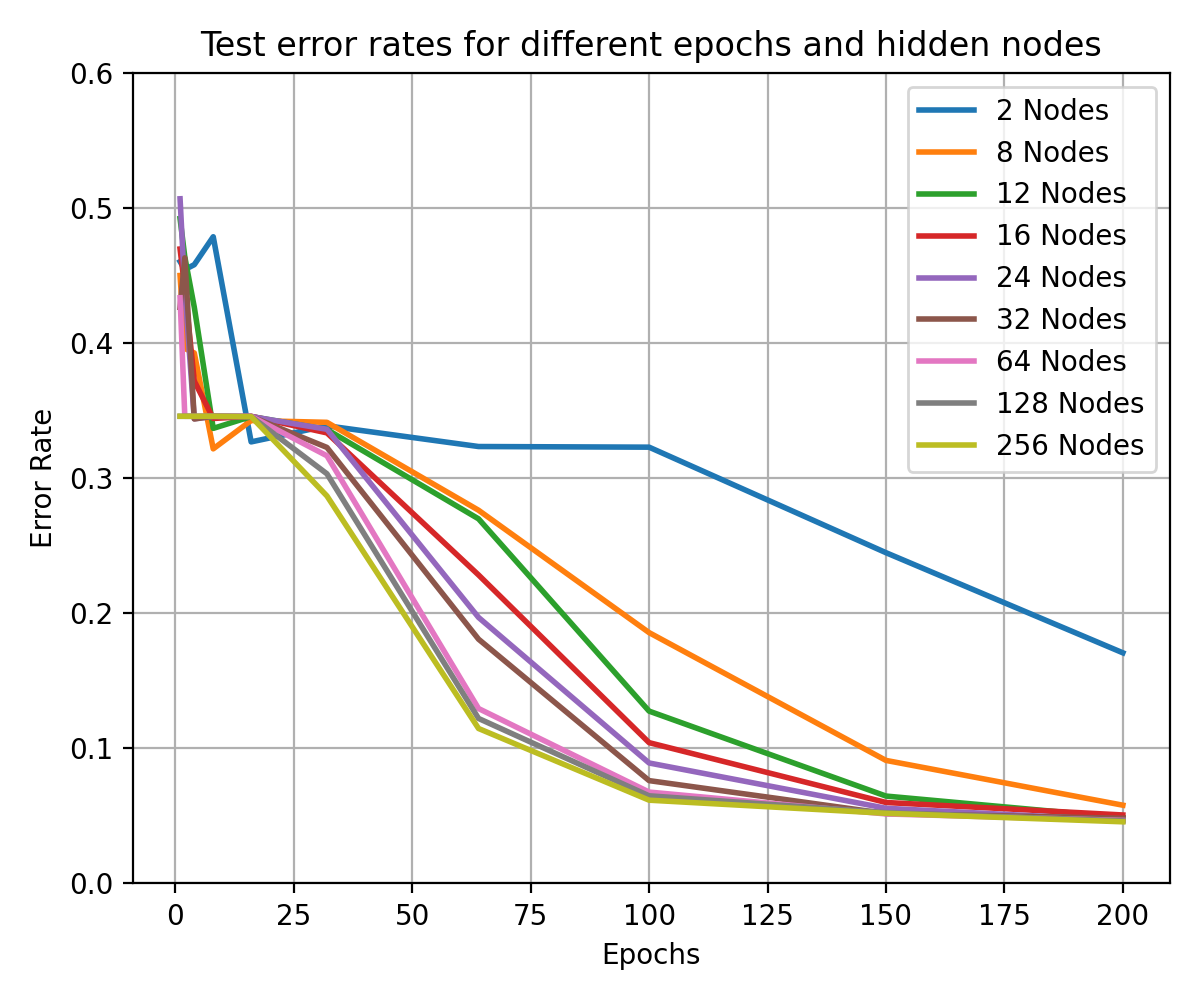
(image error) Size: 166 KiB 

|
|
Before 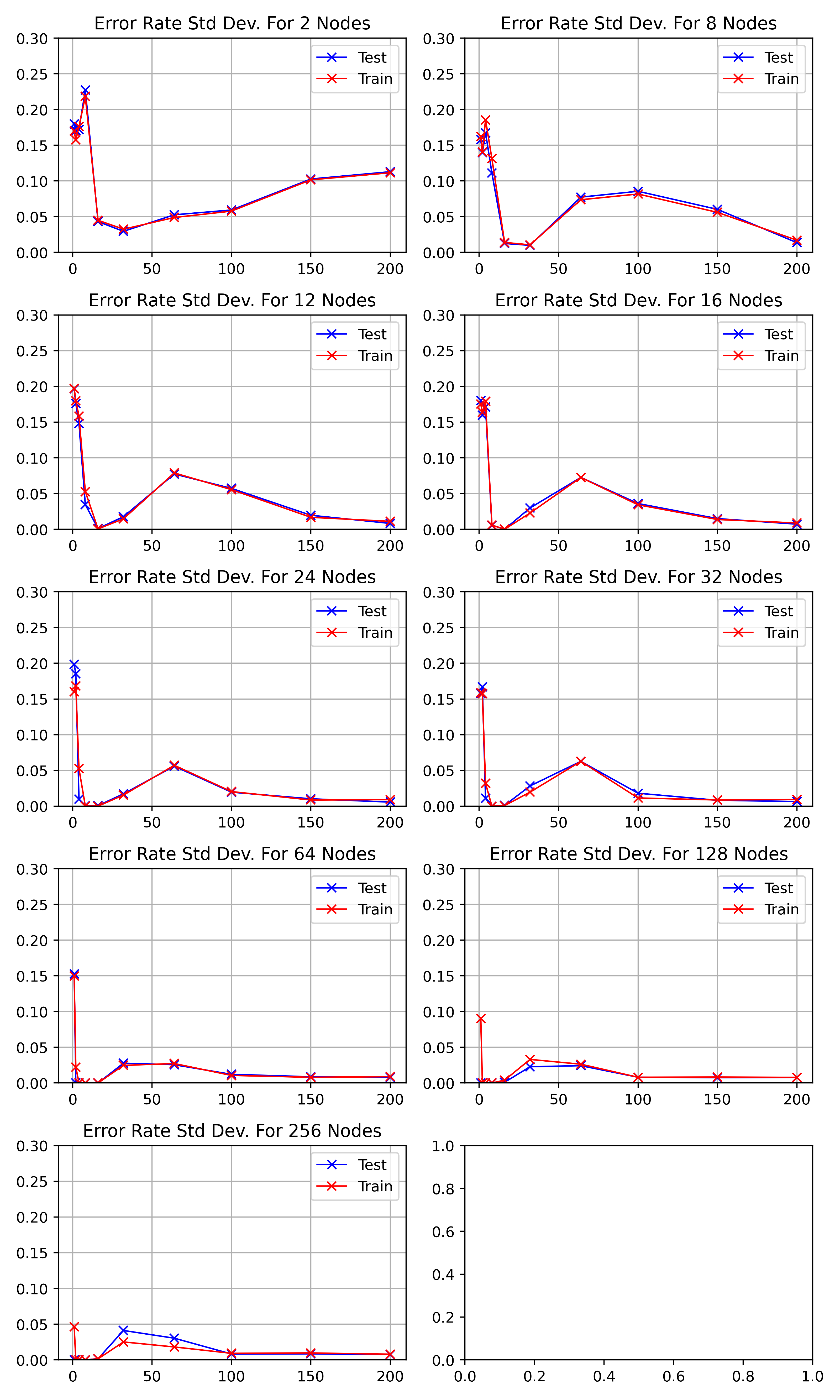
(image error) Size: 584 KiB After 
(image error) Size: 200 KiB 

|
|
Before 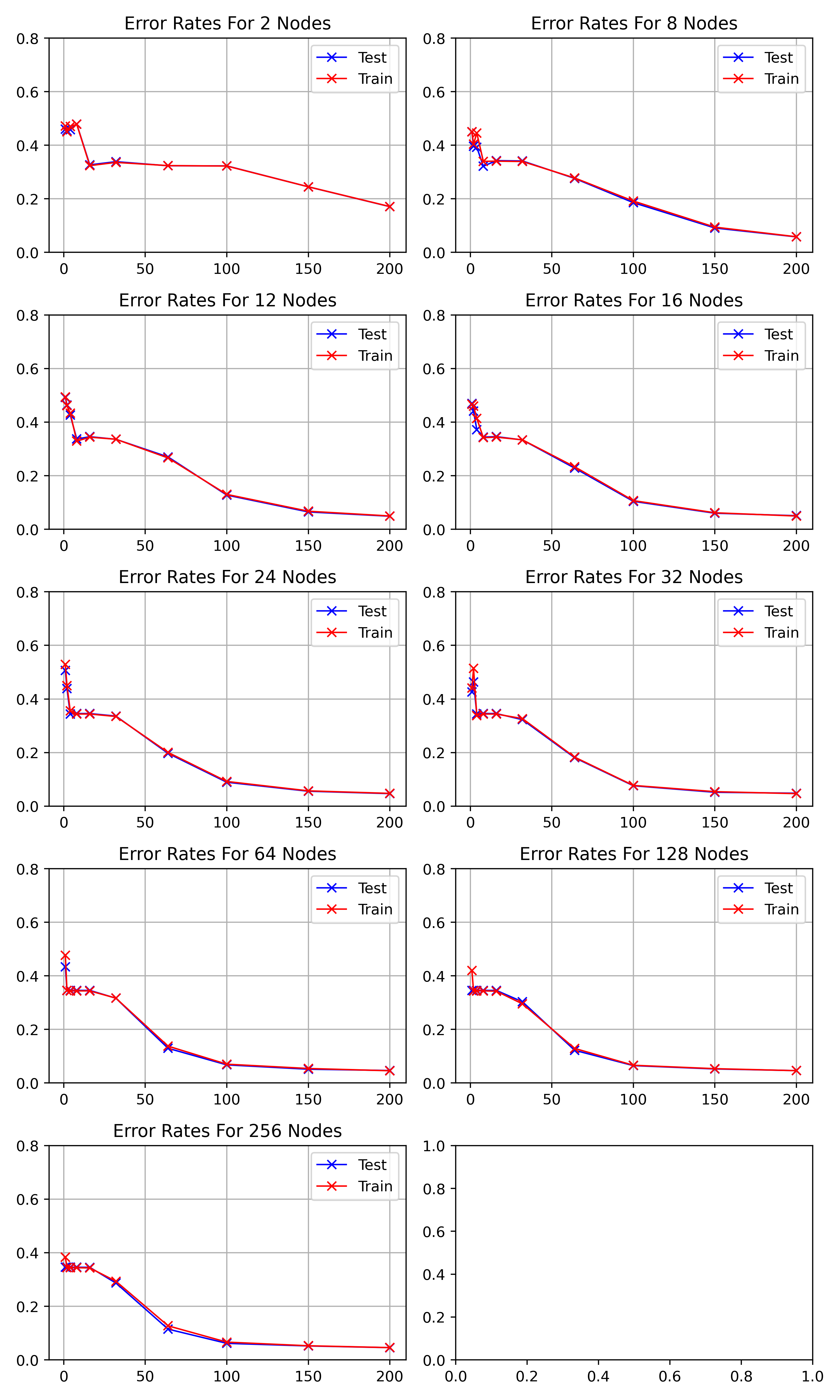
(image error) Size: 507 KiB After 
(image error) Size: 195 KiB 

|
BIN
graphs/exp1-test2-1-error-rate-curves.png
Normal file
|
After 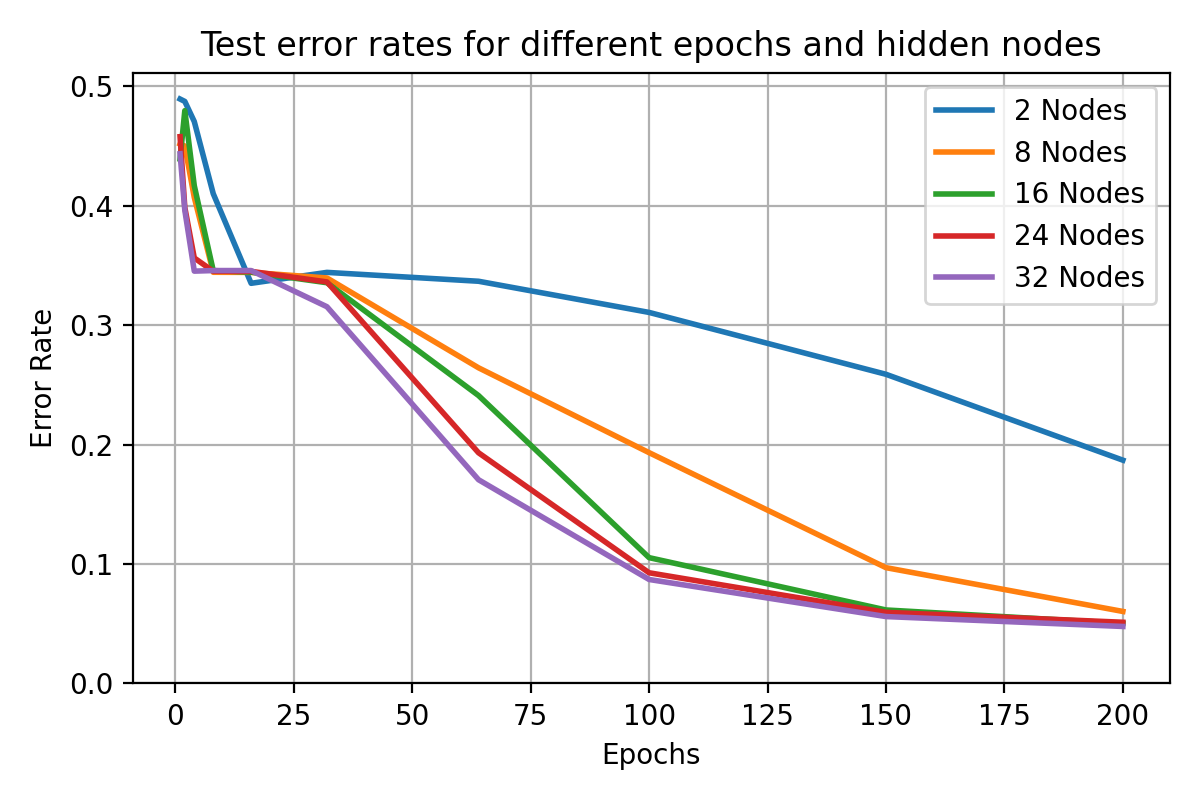
(image error) Size: 110 KiB |
BIN
graphs/exp1-test2-1-test-train-error-rate-std.png
Normal file
|
After 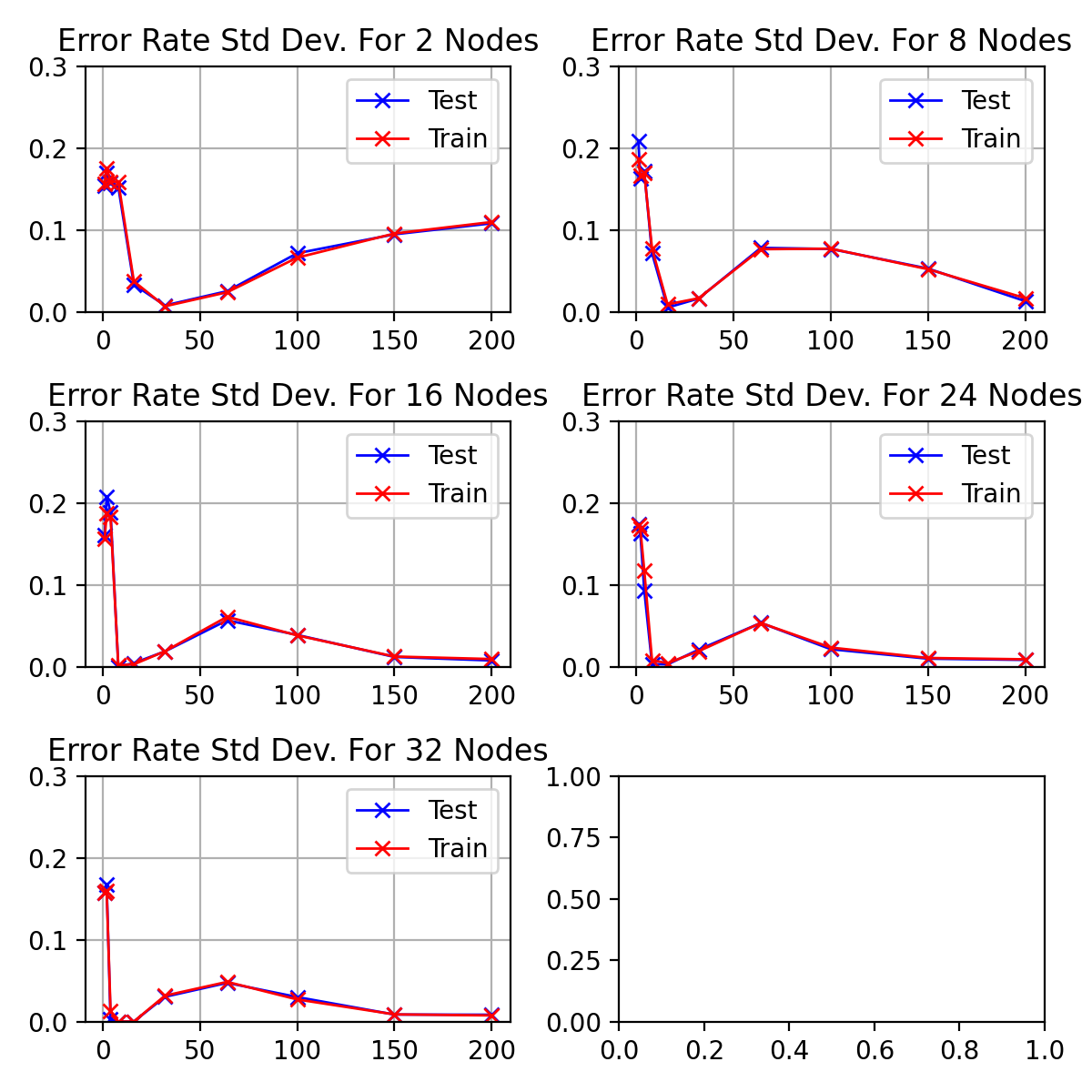
(image error) Size: 124 KiB |
BIN
graphs/exp1-test2-1-test-train-error-rate.png
Normal file
|
After 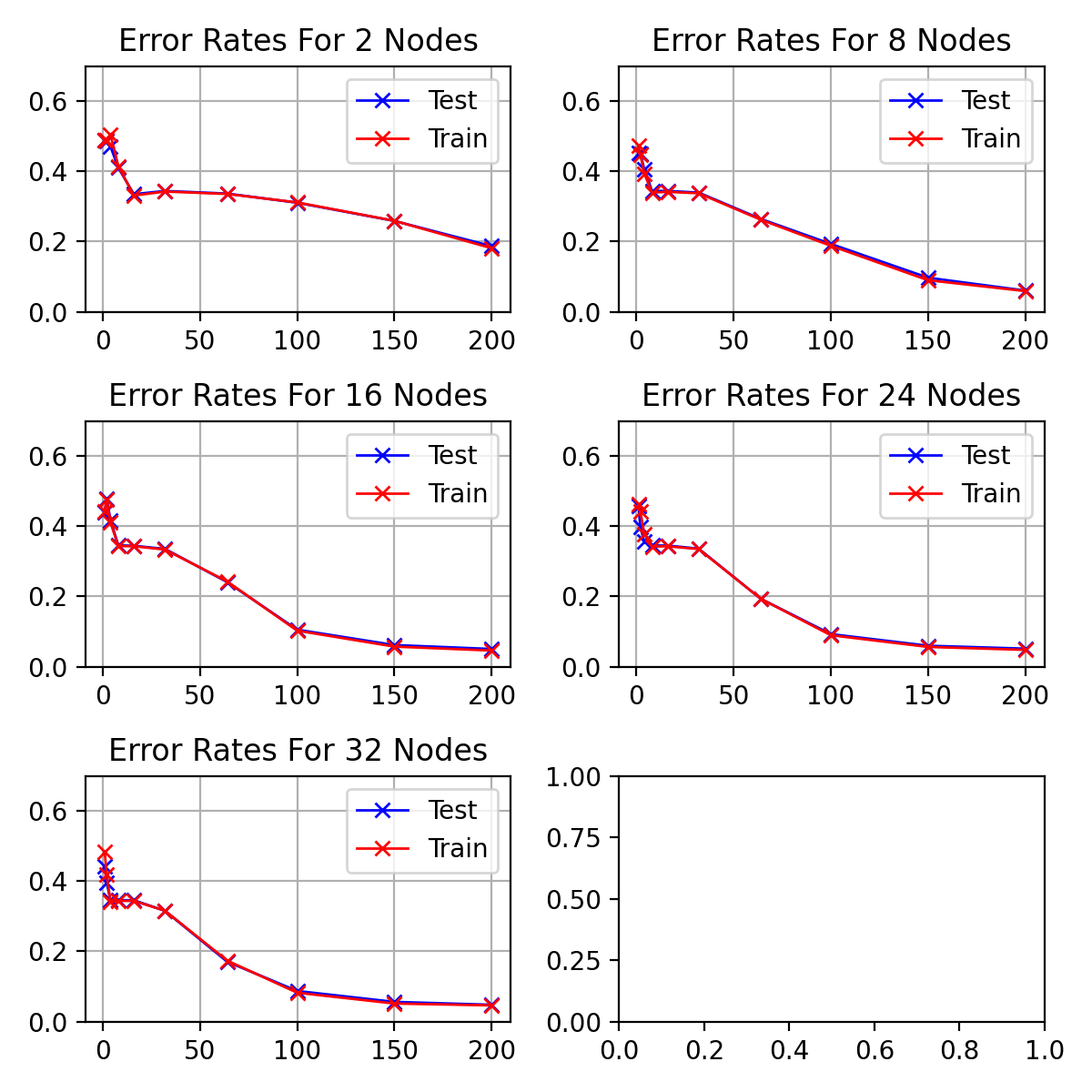
(image error) Size: 116 KiB |
BIN
graphs/exp1-test2-2-error-rate-curves.png
Normal file
|
After 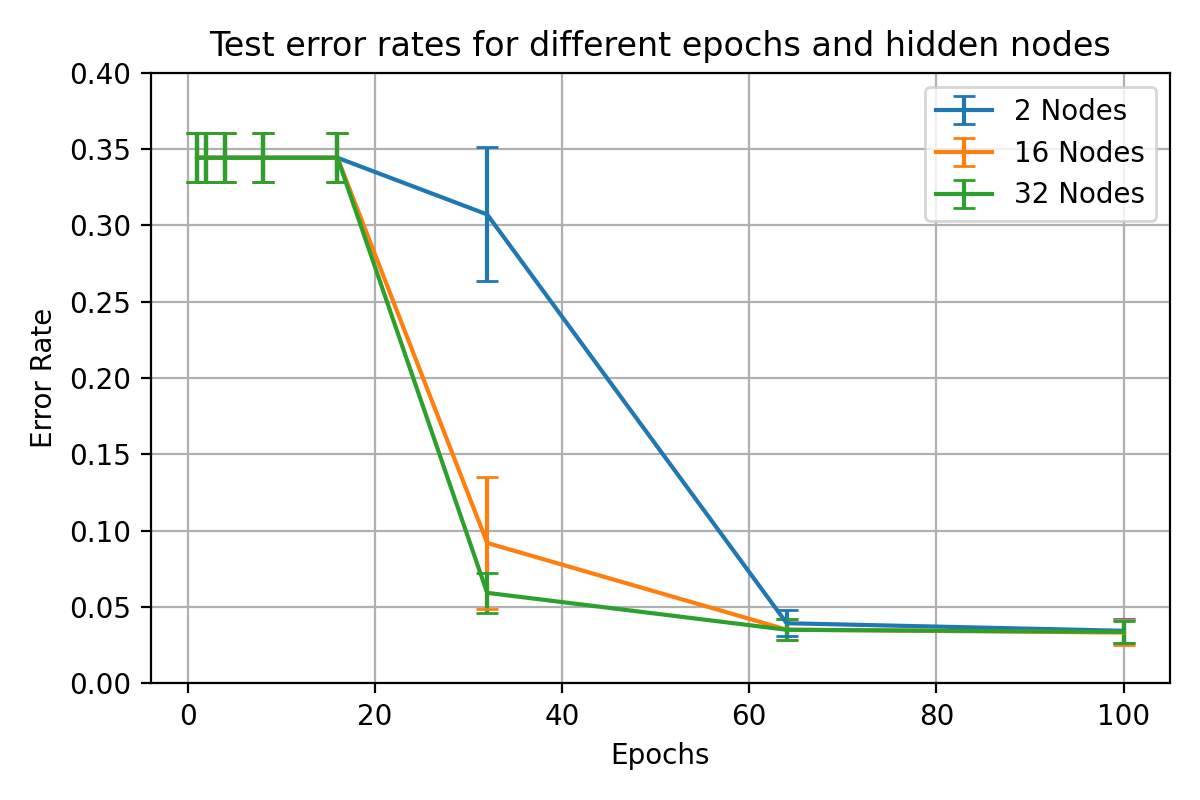
(image error) Size: 79 KiB |
BIN
graphs/exp1-test2-2-test-train-error-rate-std.png
Normal file
|
After 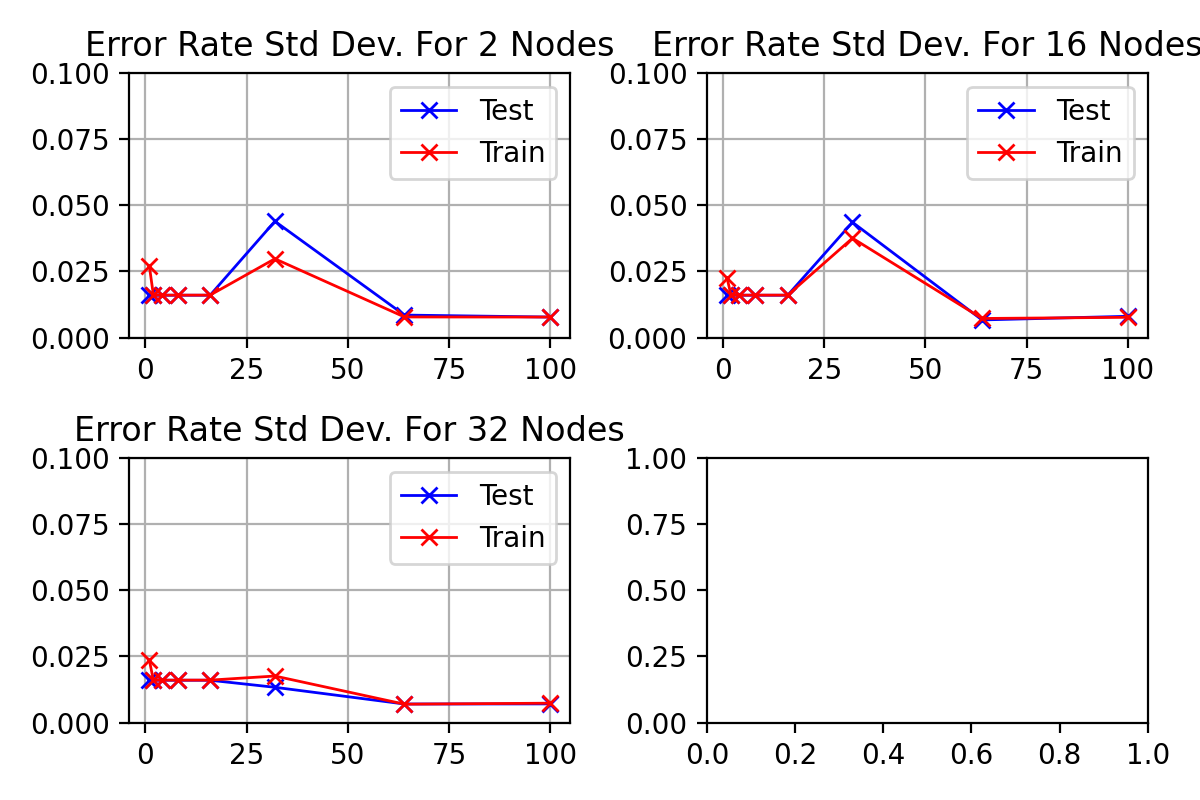
(image error) Size: 79 KiB |
BIN
graphs/exp1-test2-2-test-train-error-rate.png
Normal file
|
After 
(image error) Size: 74 KiB |
BIN
graphs/exp1-test2-3-error-rate-curves.png
Normal file
|
After 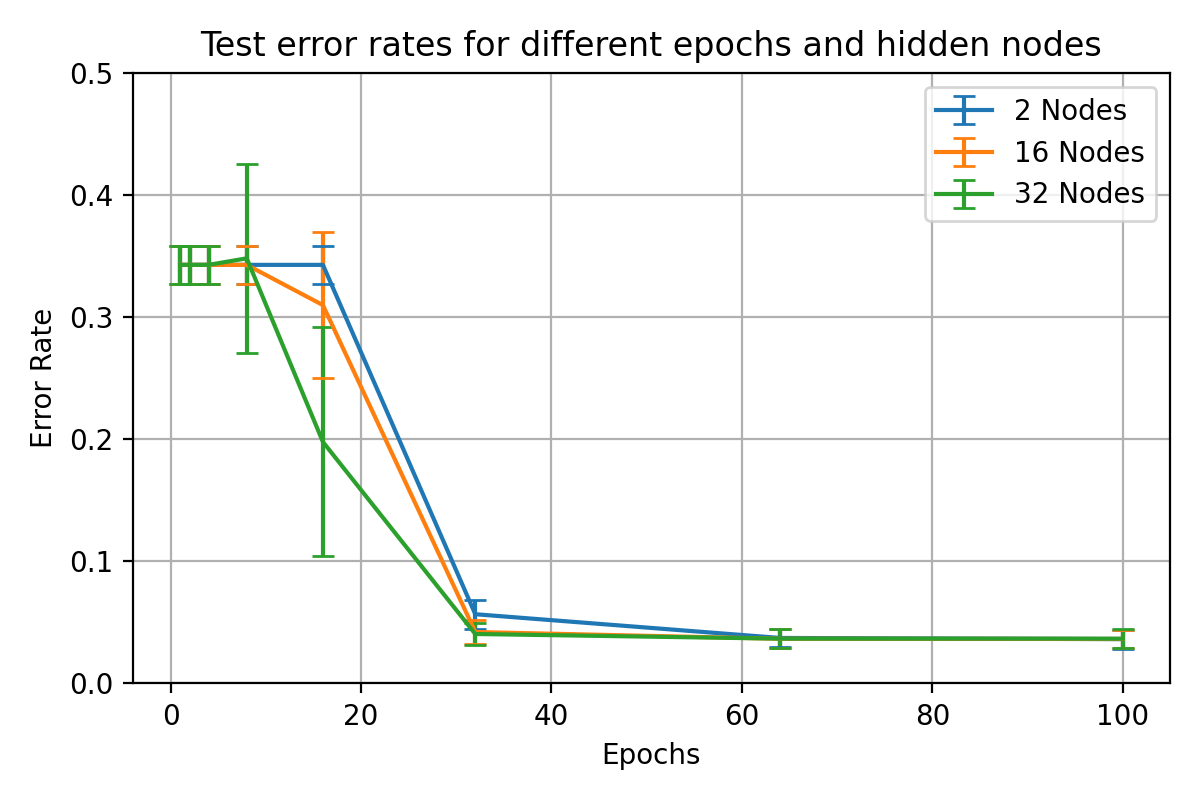
(image error) Size: 65 KiB |
BIN
graphs/exp1-test2-3-test-train-error-rate-std.png
Normal file
|
After 
(image error) Size: 81 KiB |
BIN
graphs/exp1-test2-3-test-train-error-rate.png
Normal file
|
After 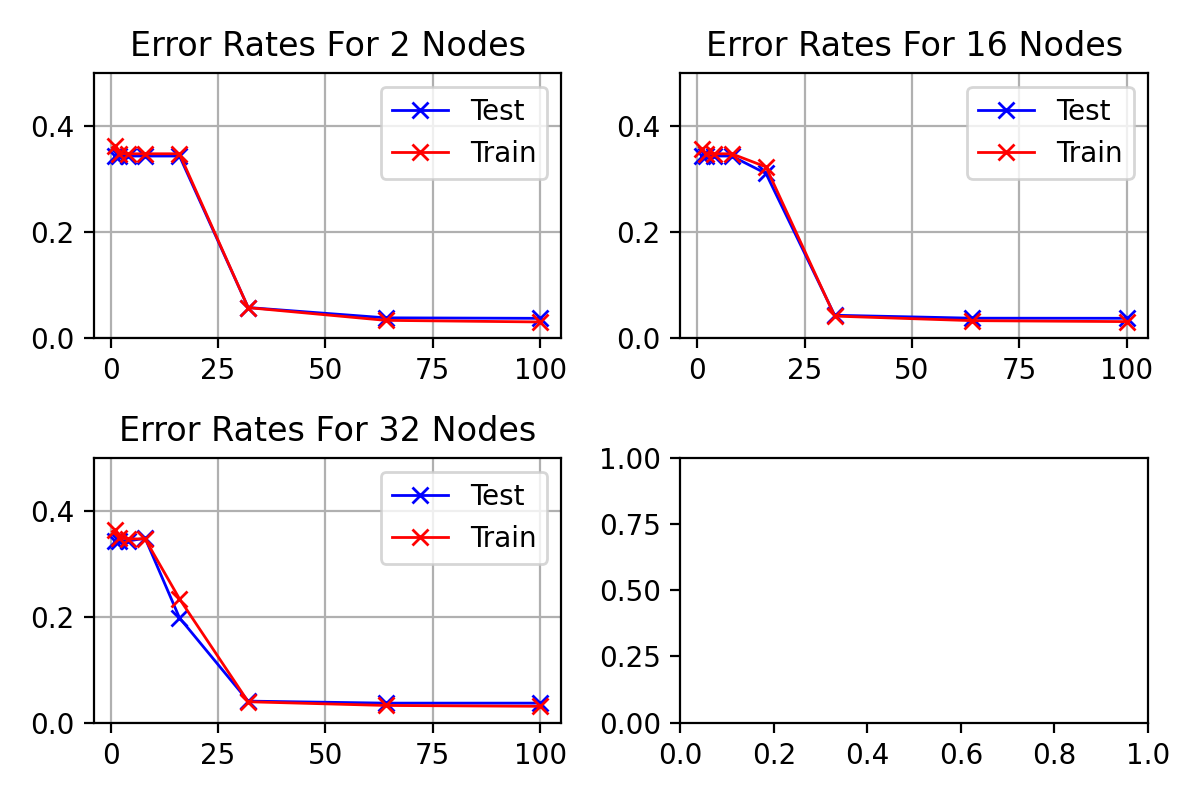
(image error) Size: 68 KiB |
|
Before 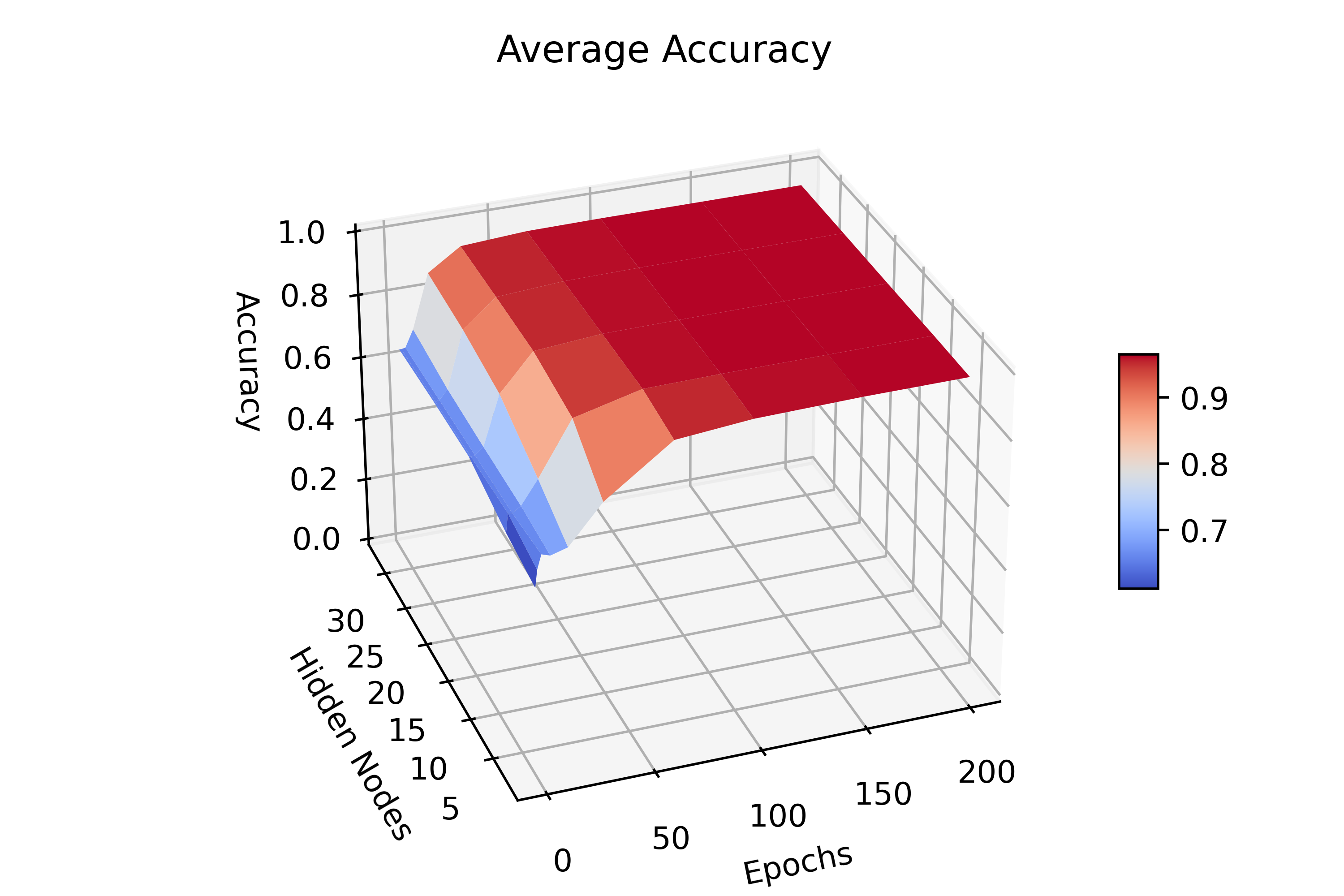
(image error) Size: 473 KiB After 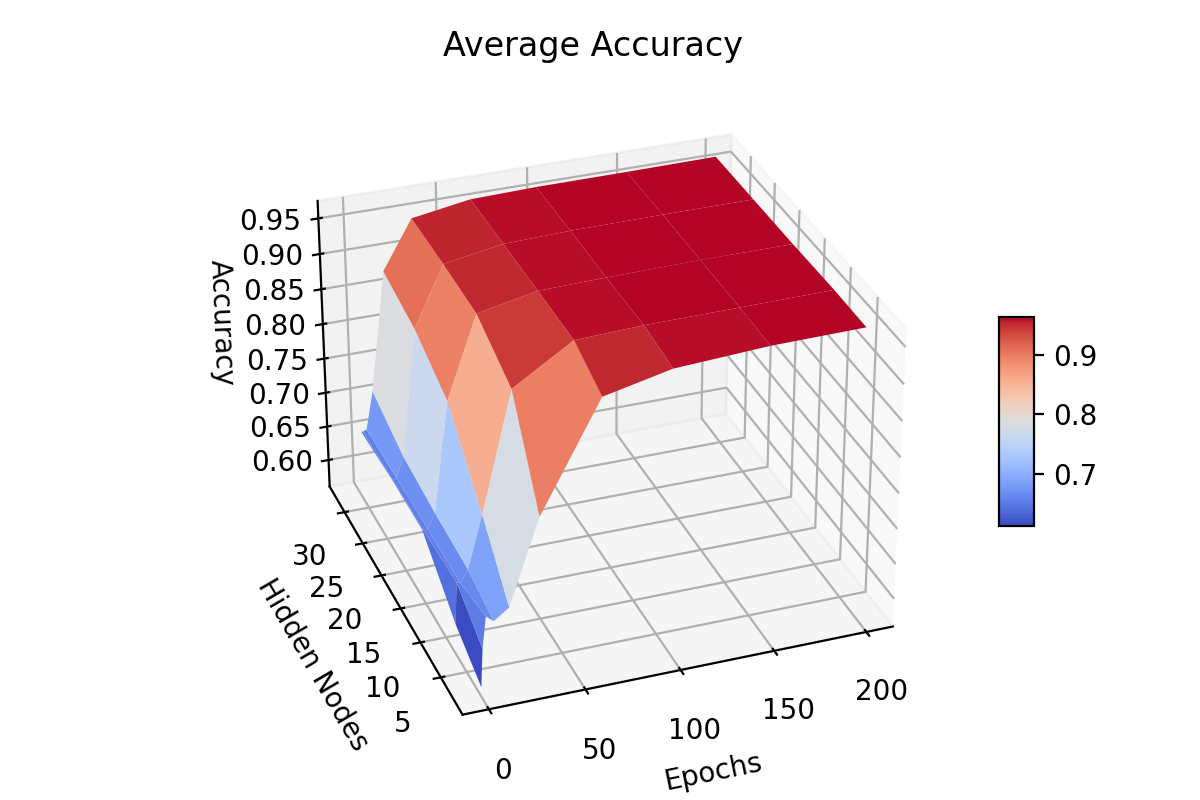
(image error) Size: 171 KiB 

|
|
Before 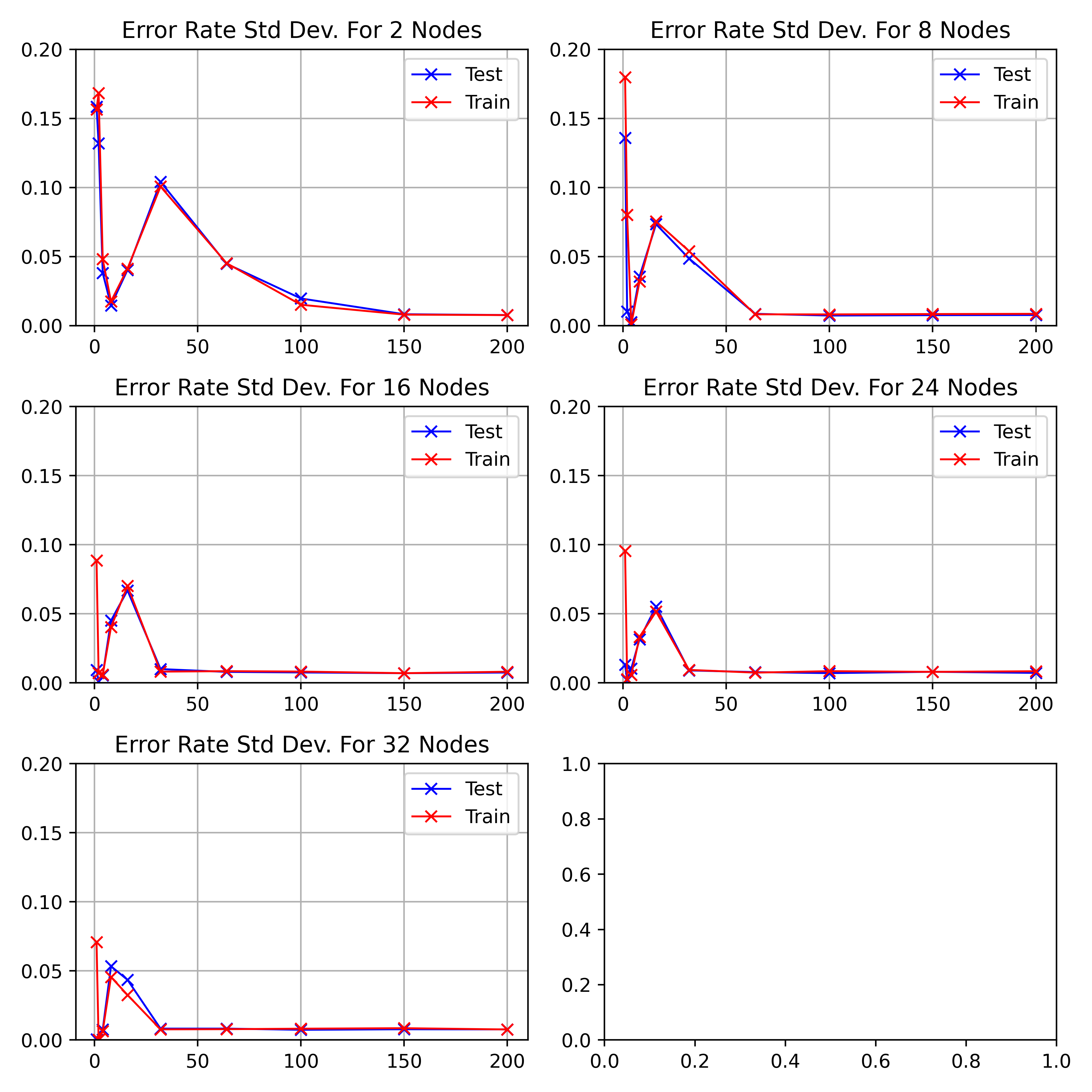
(image error) Size: 396 KiB After 
(image error) Size: 144 KiB 

|
|
Before 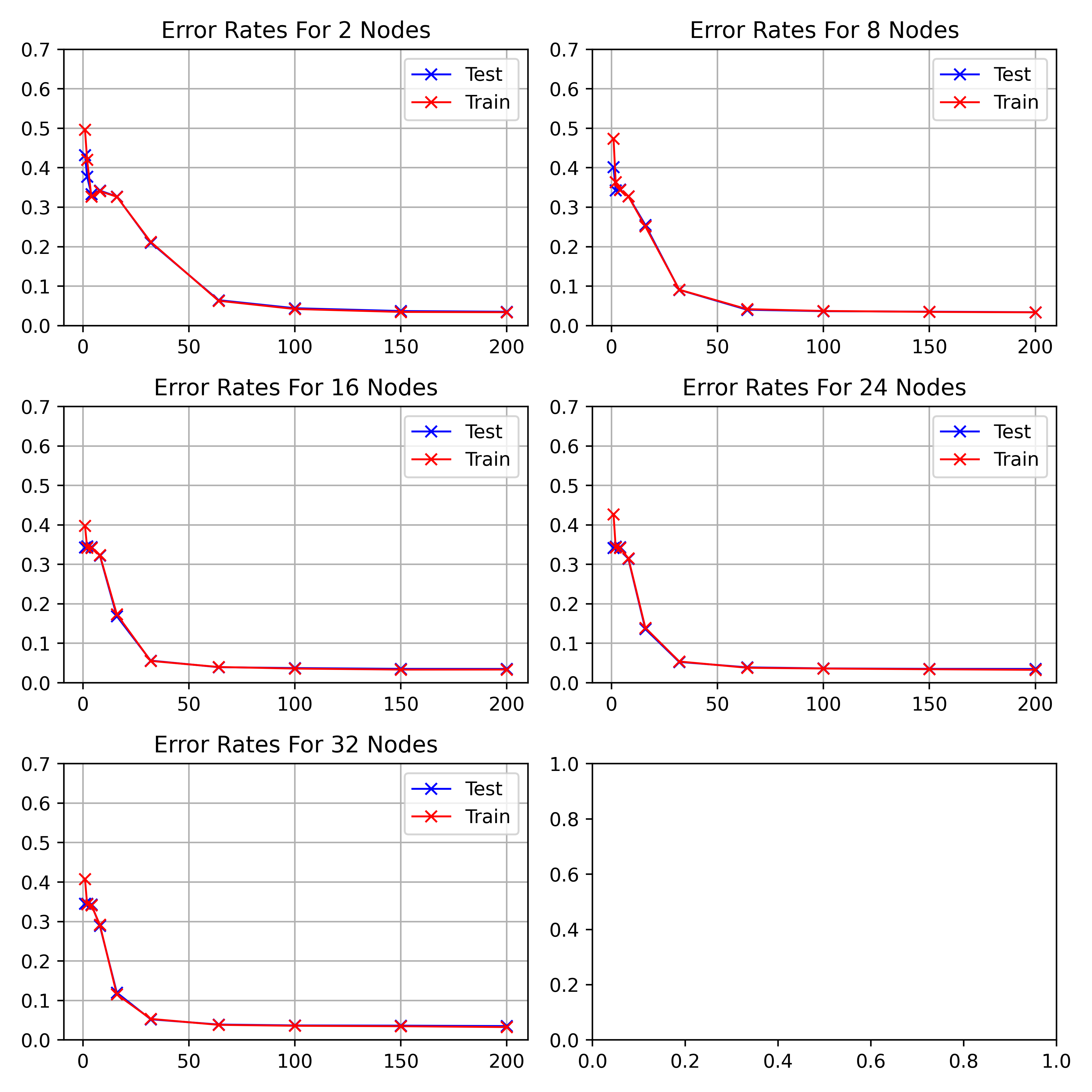
(image error) Size: 352 KiB After 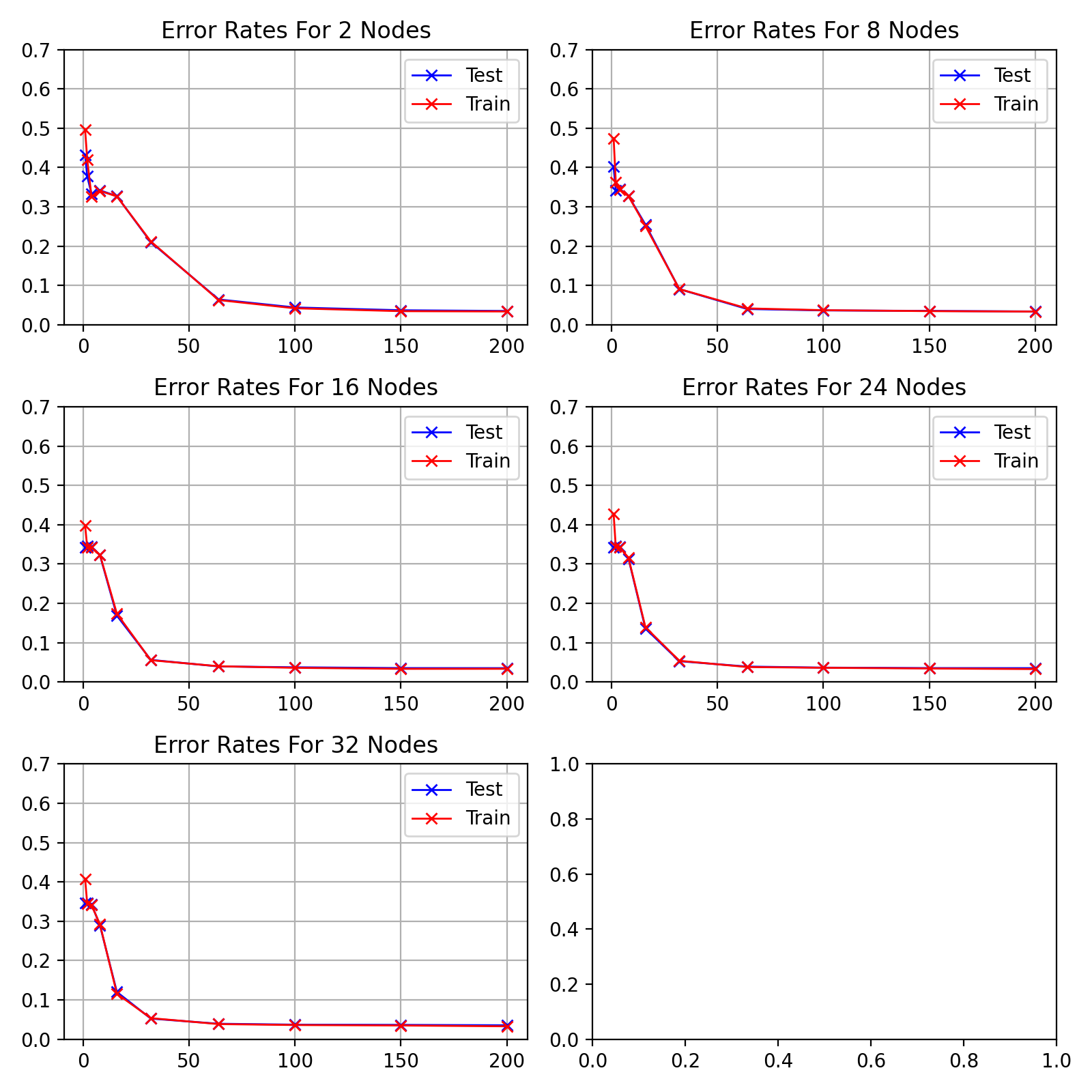
(image error) Size: 133 KiB 

|
|
Before 
(image error) Size: 474 KiB After 
(image error) Size: 170 KiB 

|
|
Before 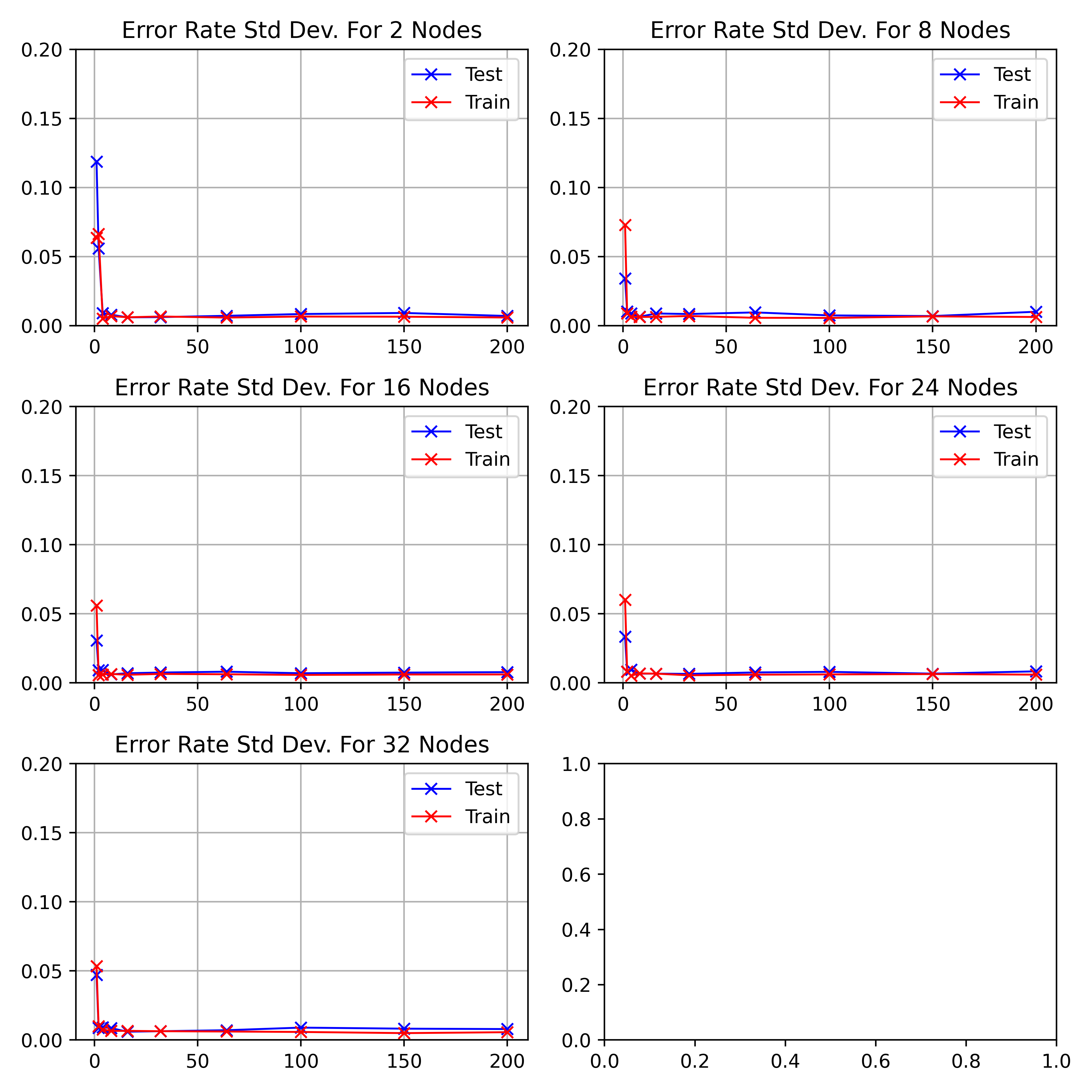
(image error) Size: 317 KiB After 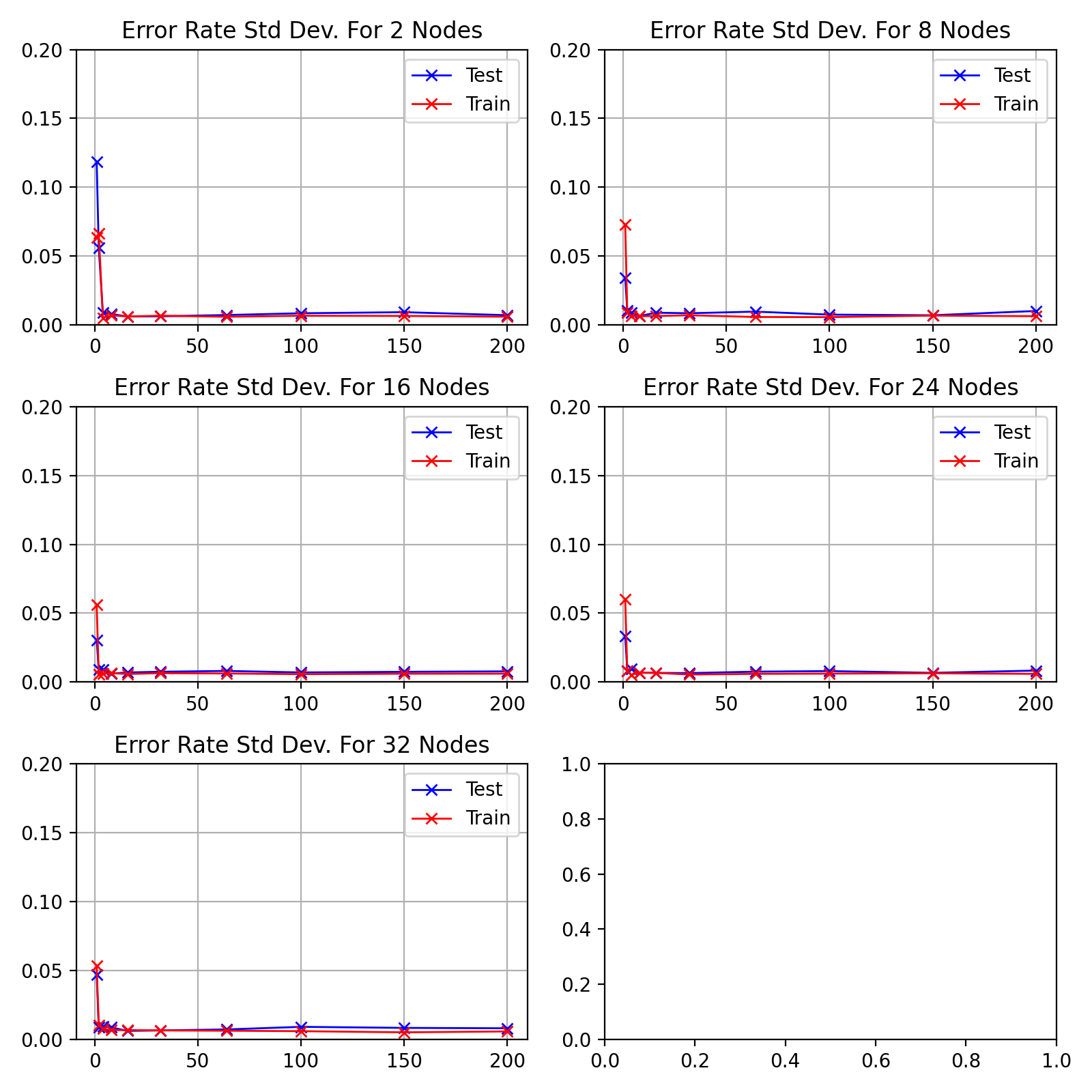
(image error) Size: 118 KiB 

|
|
Before 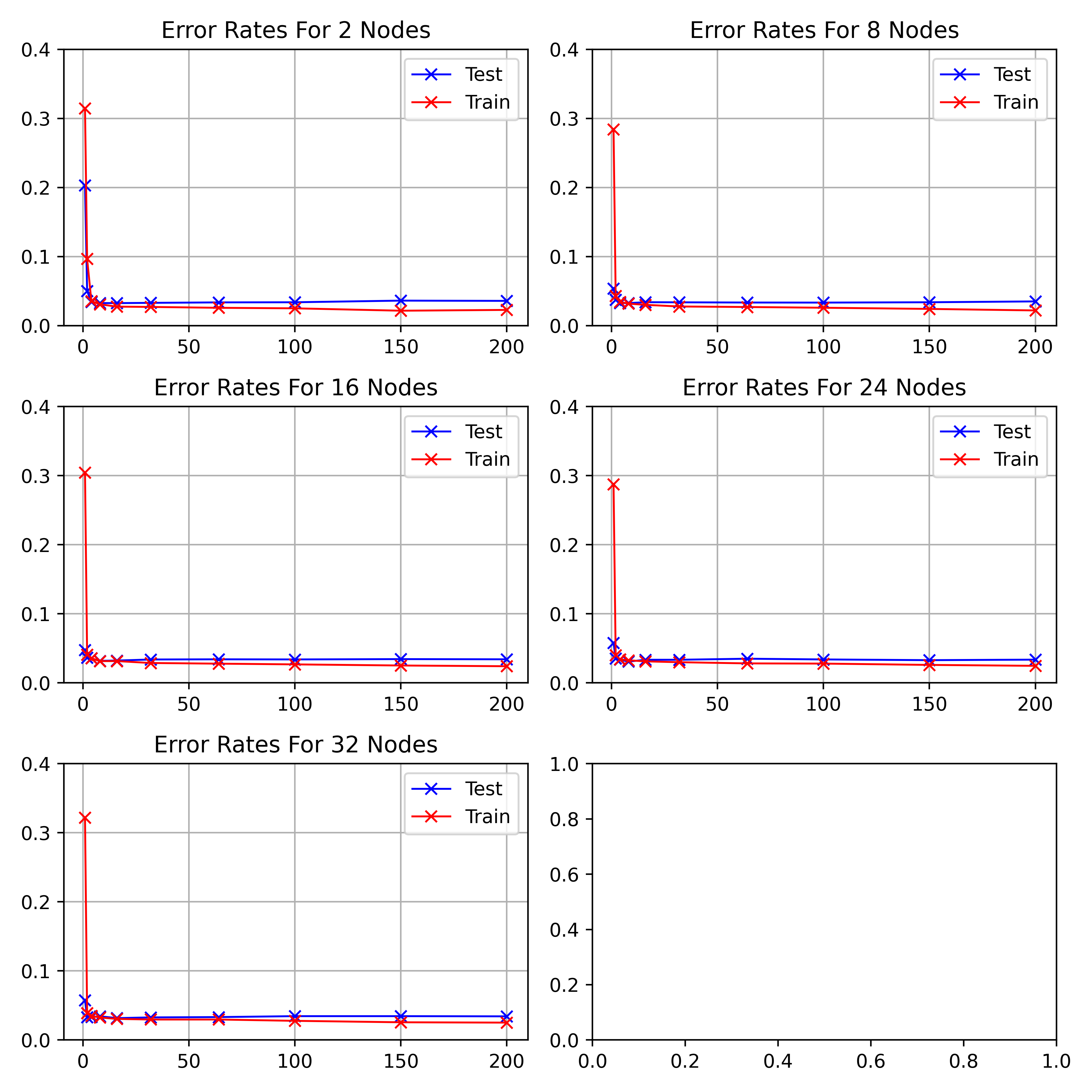
(image error) Size: 295 KiB After 
(image error) Size: 110 KiB 

|
|
Before 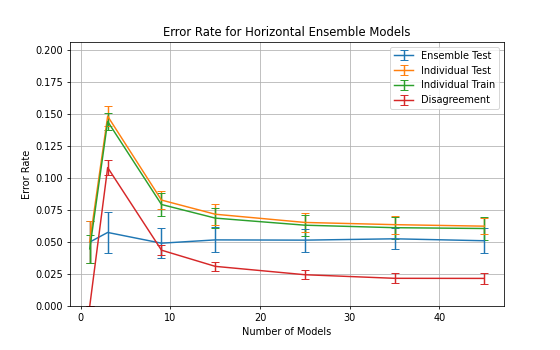
(image error) Size: 29 KiB After 
(image error) Size: 90 KiB 

|
BIN
graphs/exp2-test12-error-rate-curves.png
Normal file
|
After 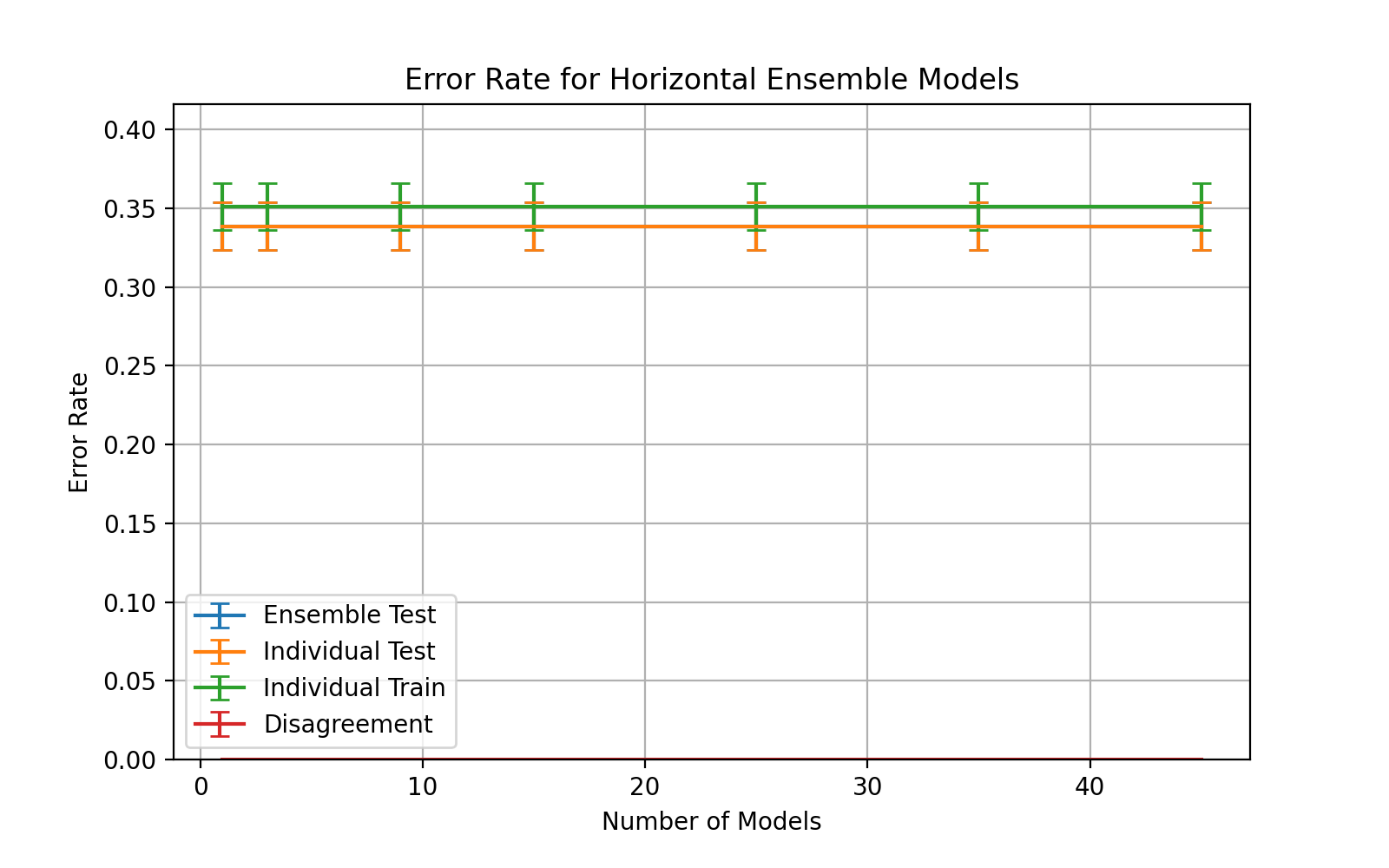
(image error) Size: 56 KiB |
BIN
graphs/exp2-test13-error-rate-curves.png
Normal file
|
After 
(image error) Size: 52 KiB |
BIN
graphs/exp2-test14-error-rate-curves.png
Normal file
|
After 
(image error) Size: 51 KiB |
BIN
graphs/exp2-test15-error-rate-curves.png
Normal file
|
After 
(image error) Size: 55 KiB |
BIN
graphs/exp2-test16-error-rate-curves.png
Normal file
|
After 
(image error) Size: 52 KiB |
BIN
graphs/exp2-test17-error-rate-curves.png
Normal file
|
After 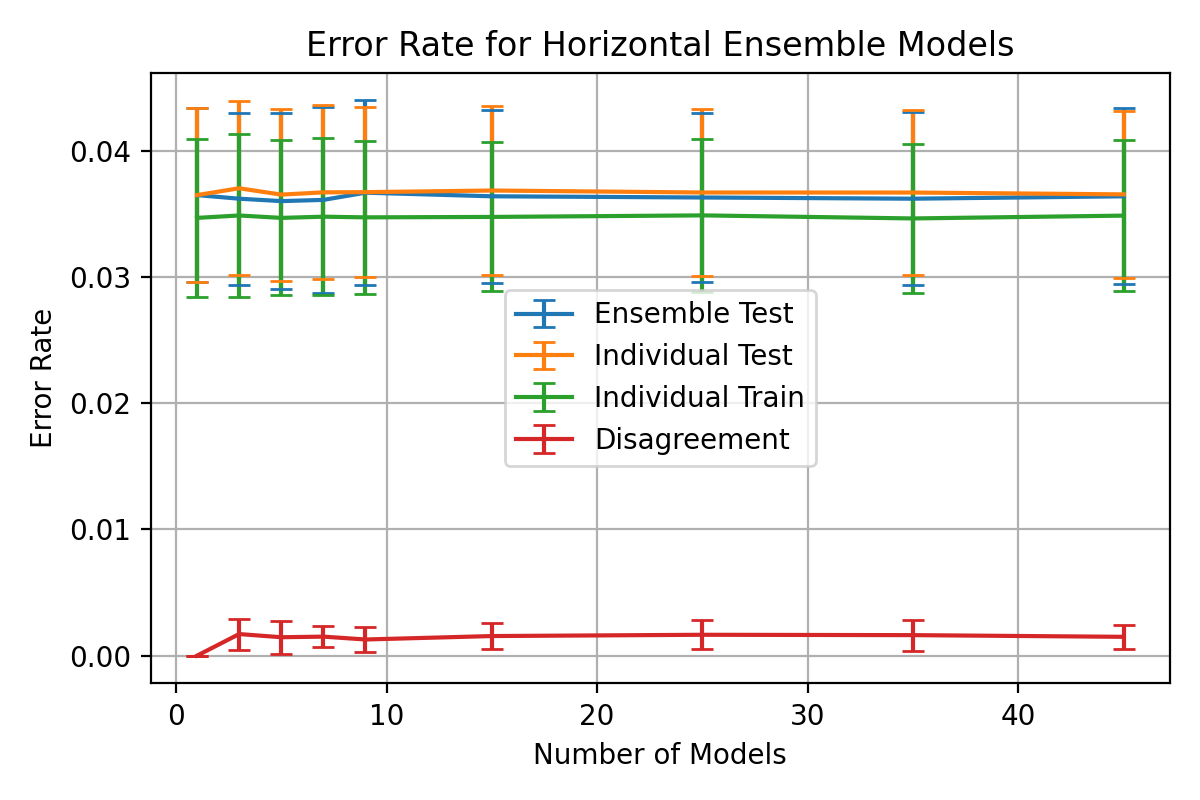
(image error) Size: 58 KiB |
|
Before 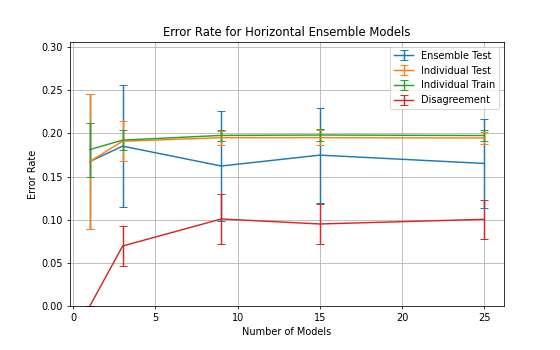
(image error) Size: 22 KiB After 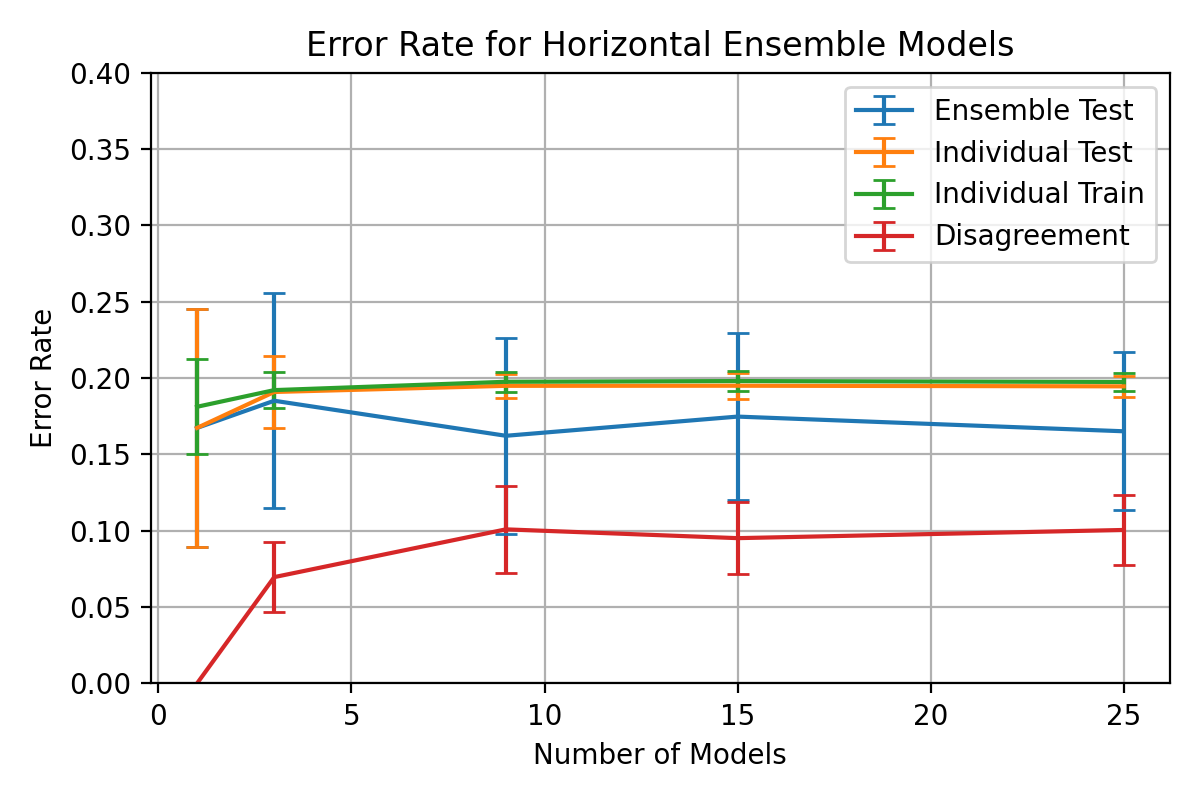
(image error) Size: 68 KiB 

|
|
Before 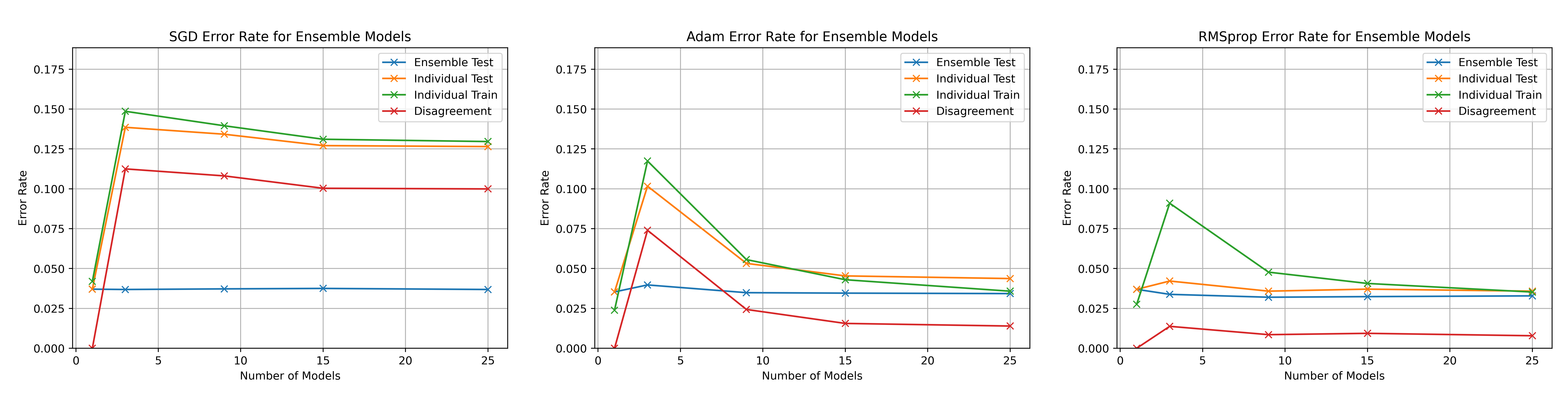
(image error) Size: 401 KiB After 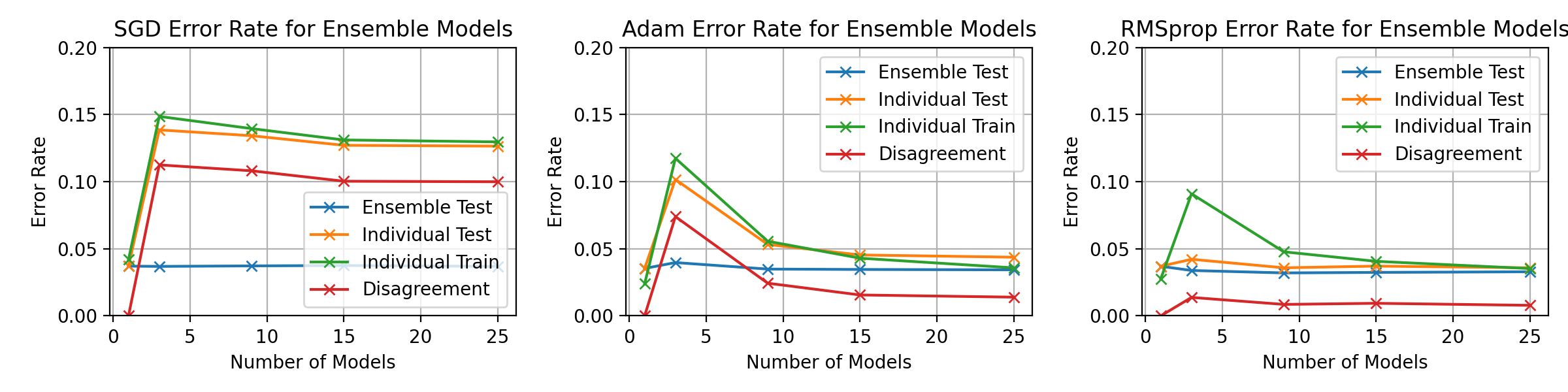
(image error) Size: 140 KiB 

|
BIN
graphs/exp3-test2-error-rate-curves.png
Normal file
|
After 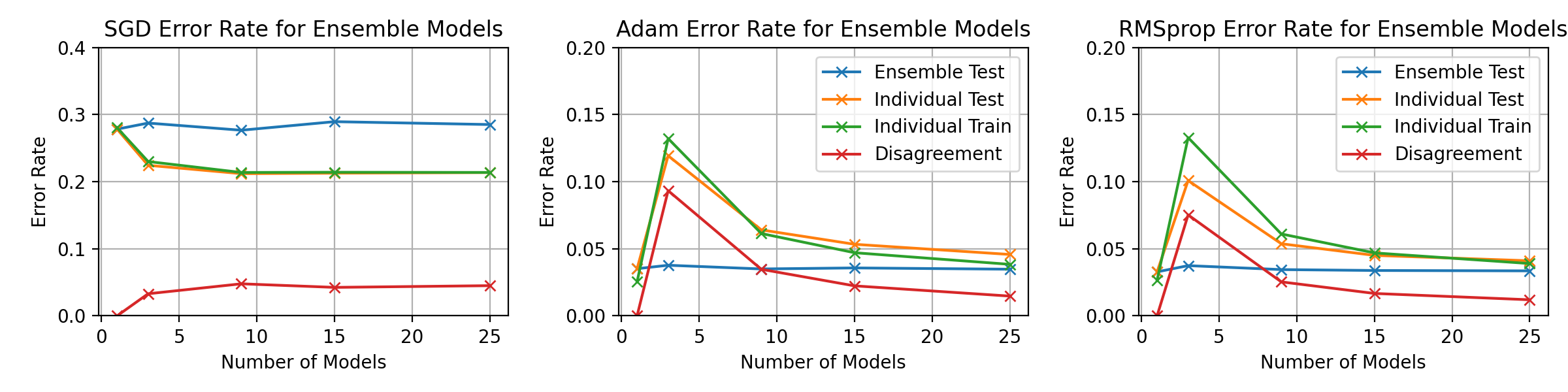
(image error) Size: 130 KiB |
BIN
graphs/exp3-test3-error-rate-curves.png
Normal file
|
After 
(image error) Size: 110 KiB |
BIN
graphs/exp3-test4-error-rate-curves.png
Normal file
|
After 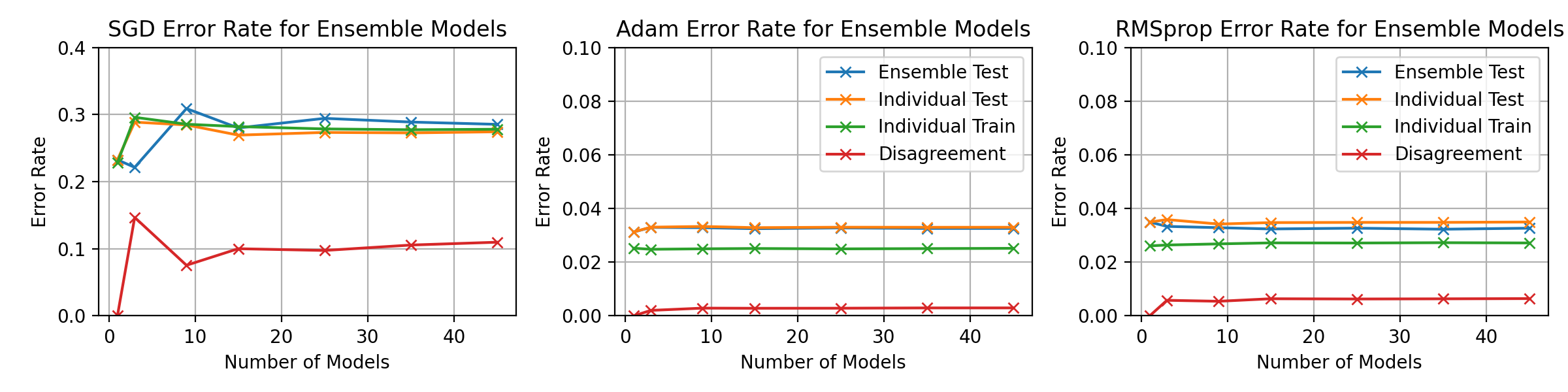
(image error) Size: 106 KiB |
BIN
graphs/exp3-test5-error-rate-curves.png
Normal file
|
After 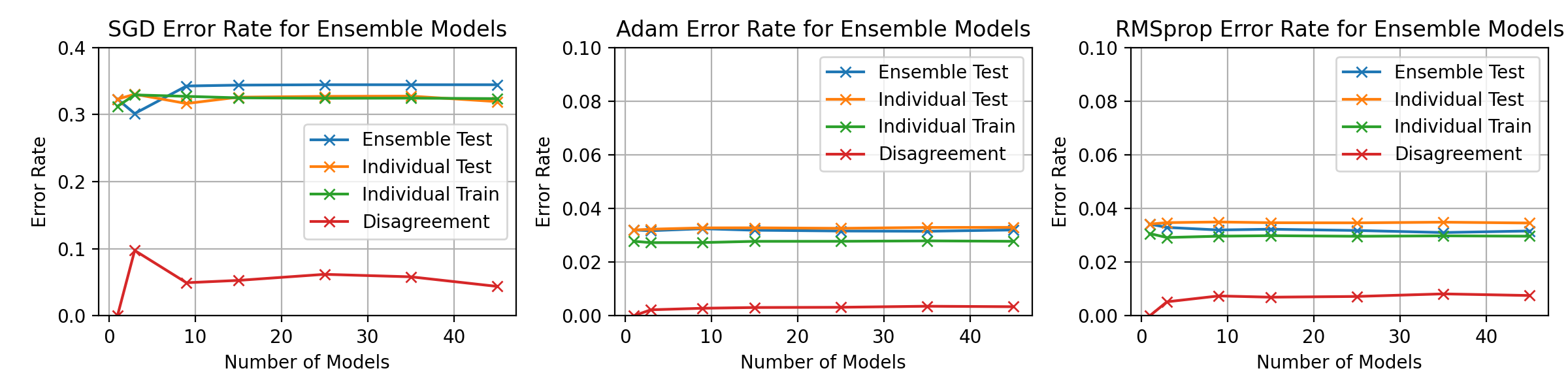
(image error) Size: 119 KiB |
BIN
graphs/exp3-test6-error-rate-curves.png
Normal file
|
After 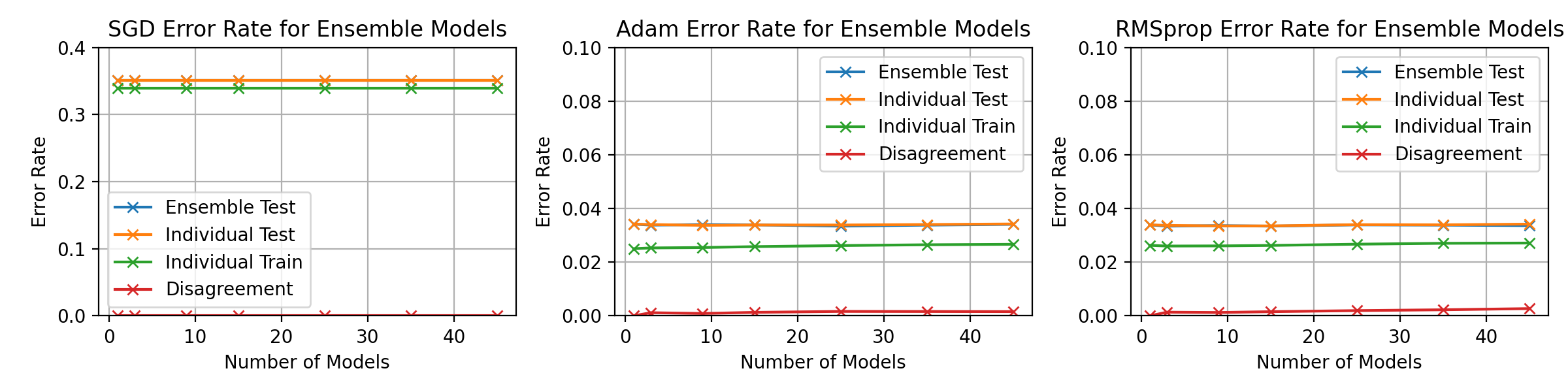
(image error) Size: 100 KiB |
BIN
graphs/exp3-test7-error-rate-curves.png
Normal file
|
After 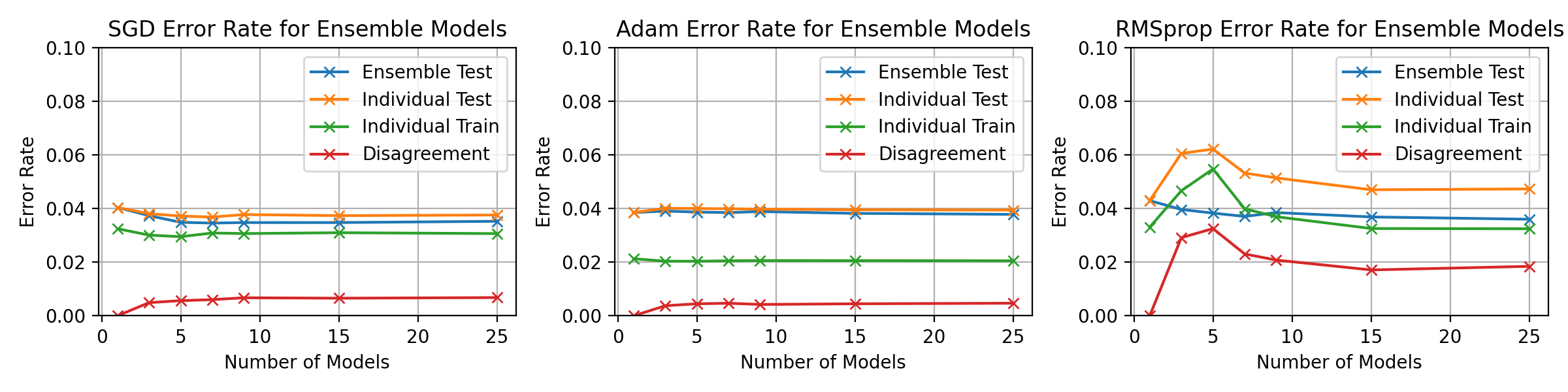
(image error) Size: 110 KiB |
BIN
graphs/exp3-test8-error-rate-curves.png
Normal file
|
After 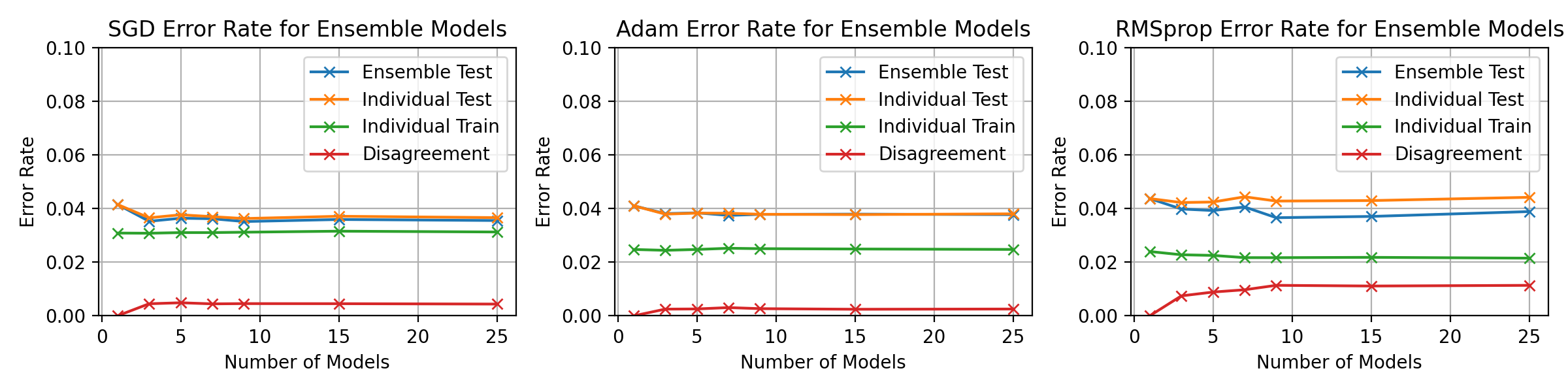
(image error) Size: 99 KiB |
285
nncw.ipynb
@ -404,7 +404,7 @@ noprefix "false"
|
||||
in conjunction.
|
||||
The effect of varying the number of nodes and epochs throughout the ensemble
|
||||
was considered in order to determine whether combining multiple models
|
||||
could produce a better accuracy than those individually.
|
||||
could produce a better accuracy than any individual model.
|
||||
Section
|
||||
\begin_inset CommandInset ref
|
||||
LatexCommand ref
|
||||
@ -432,7 +432,7 @@ noprefix "false"
|
||||
\end_layout
|
||||
|
||||
\begin_layout Section
|
||||
Hidden Nodes & Epochs (Exp 1)
|
||||
Hidden Nodes & Epochs
|
||||
\begin_inset CommandInset label
|
||||
LatexCommand label
|
||||
name "sec:exp1"
|
||||
@ -443,21 +443,257 @@ name "sec:exp1"
|
||||
\end_layout
|
||||
|
||||
\begin_layout Standard
|
||||
This section investigates the effect of varying the number of hidden nodes
|
||||
in a single hidden layer of a multi-layer perceptron.
|
||||
This is compared to the effect of varying
|
||||
This section investigates the effect of varying the number of nodes in the
|
||||
single hidden layer of a shallow multi-layer perceptron.
|
||||
This is compared to the effect of training the model with different numbers
|
||||
of epochs.
|
||||
Throughout the experiment, stochastic gradient descent with momentum is
|
||||
used as the optimiser, variations in both momentum and learning rate are
|
||||
presented.
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Subsection
|
||||
Results
|
||||
\end_layout
|
||||
|
||||
\begin_layout Standard
|
||||
\begin_inset Float figure
|
||||
wide false
|
||||
sideways false
|
||||
status open
|
||||
|
||||
\begin_layout Plain Layout
|
||||
\noindent
|
||||
\align center
|
||||
\begin_inset Graphics
|
||||
filename ../graphs/exp1-test1-error-rate-curves.png
|
||||
lyxscale 50
|
||||
width 50col%
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Plain Layout
|
||||
\begin_inset Caption Standard
|
||||
|
||||
\begin_layout Plain Layout
|
||||
Varied hidden node performance results over varied training lengths for
|
||||
|
||||
\begin_inset Formula $\eta=0.01$
|
||||
\end_inset
|
||||
|
||||
,
|
||||
\begin_inset Formula $p=0$
|
||||
\end_inset
|
||||
|
||||
|
||||
\begin_inset CommandInset label
|
||||
LatexCommand label
|
||||
name "fig:exp1-test1"
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Plain Layout
|
||||
|
||||
\end_layout
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Standard
|
||||
Figure
|
||||
\begin_inset CommandInset ref
|
||||
LatexCommand ref
|
||||
reference "fig:exp1-test1"
|
||||
plural "false"
|
||||
caps "false"
|
||||
noprefix "false"
|
||||
|
||||
\end_inset
|
||||
|
||||
visualises the performance of hidden nodes up to 256 over training periods
|
||||
up to 200 epochs in length.
|
||||
In general, the error rate can be seen to decrease when the models are
|
||||
trained for longer.
|
||||
Increasing the number of nodes decreases the error rate and increases the
|
||||
gradient with which it falls up to a limit.
|
||||
64, 128 and 256 hidden nodes lie close together as the increases in performance
|
||||
slow.
|
||||
Between 0 and 25 epochs, the error rate throughout for any number of nodes
|
||||
can descend little below 0.35.
|
||||
The number of epochs to overcome this plateau is different for each number
|
||||
of nodes.
|
||||
\end_layout
|
||||
|
||||
\begin_layout Standard
|
||||
The standard deviations for the above discussed results of figure
|
||||
\begin_inset CommandInset ref
|
||||
LatexCommand ref
|
||||
reference "fig:exp1-test1"
|
||||
plural "false"
|
||||
caps "false"
|
||||
noprefix "false"
|
||||
|
||||
\end_inset
|
||||
|
||||
can be seen in figure
|
||||
\begin_inset CommandInset ref
|
||||
LatexCommand ref
|
||||
reference "fig:exp1-test1-std"
|
||||
plural "false"
|
||||
caps "false"
|
||||
noprefix "false"
|
||||
|
||||
\end_inset
|
||||
|
||||
.
|
||||
As the network starts training, the standard deviation decreases to a minimum
|
||||
between
|
||||
\begin_inset Formula $10-20$
|
||||
\end_inset
|
||||
|
||||
epochs before increasing to a peak at 64.
|
||||
As the number of hidden nodes increases, the standard deviation decreases.
|
||||
The initial drop is sharper and the 64 epoch peak increases higher.
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Standard
|
||||
\begin_inset Float figure
|
||||
wide false
|
||||
sideways false
|
||||
status open
|
||||
|
||||
\begin_layout Plain Layout
|
||||
\noindent
|
||||
\align center
|
||||
\begin_inset Graphics
|
||||
filename /mnt/files/dev/py/shallow-training/graphs/exp1-test1-test-train-error-rate-std.png
|
||||
lyxscale 50
|
||||
width 60col%
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Plain Layout
|
||||
\begin_inset Caption Standard
|
||||
|
||||
\begin_layout Plain Layout
|
||||
Standard deviation of results from figure
|
||||
\begin_inset CommandInset ref
|
||||
LatexCommand ref
|
||||
reference "fig:exp1-test1"
|
||||
plural "false"
|
||||
caps "false"
|
||||
noprefix "false"
|
||||
|
||||
\end_inset
|
||||
|
||||
with
|
||||
\begin_inset Formula $\eta=0.01$
|
||||
\end_inset
|
||||
|
||||
,
|
||||
\begin_inset Formula $p=0$
|
||||
\end_inset
|
||||
|
||||
|
||||
\begin_inset CommandInset label
|
||||
LatexCommand label
|
||||
name "fig:exp1-test1-std"
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Plain Layout
|
||||
|
||||
\end_layout
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Standard
|
||||
\begin_inset Float figure
|
||||
wide false
|
||||
sideways false
|
||||
status open
|
||||
|
||||
\begin_layout Plain Layout
|
||||
\noindent
|
||||
\align center
|
||||
\begin_inset Graphics
|
||||
filename /mnt/files/dev/py/shallow-training/graphs/exp1-test2-2-error-rate-curves.png
|
||||
lyxscale 50
|
||||
width 50col%
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Plain Layout
|
||||
\begin_inset Caption Standard
|
||||
|
||||
\begin_layout Plain Layout
|
||||
Varied hidden node performance results over varied training lengths for
|
||||
|
||||
\begin_inset Formula $\eta=0.1$
|
||||
\end_inset
|
||||
|
||||
,
|
||||
\begin_inset Formula $p=0$
|
||||
\end_inset
|
||||
|
||||
|
||||
\begin_inset CommandInset label
|
||||
LatexCommand label
|
||||
name "fig:exp1-test2-2"
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Subsection
|
||||
Discussion
|
||||
\end_layout
|
||||
|
||||
\begin_layout Section
|
||||
Ensemble Classification (Exp 2)
|
||||
Ensemble Classification
|
||||
\begin_inset CommandInset label
|
||||
LatexCommand label
|
||||
name "sec:exp2"
|
||||
@ -467,16 +703,239 @@ name "sec:exp2"
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Standard
|
||||
A horizontal ensemble of
|
||||
\begin_inset Formula $m$
|
||||
\end_inset
|
||||
|
||||
models was constructed with majority vote in order to investigate whether
|
||||
this could improve performance over that of any single model.
|
||||
In order to introduce variation between models of the ensemble, a range
|
||||
for hidden nodes and epochs could be defined.
|
||||
When selecting parameters throughout the ensemble, the models are equally
|
||||
distributed throughout the ranges
|
||||
\begin_inset Foot
|
||||
status open
|
||||
|
||||
\begin_layout Plain Layout
|
||||
For
|
||||
\begin_inset Formula $m=1$
|
||||
\end_inset
|
||||
|
||||
, the average of the range is taken
|
||||
\end_layout
|
||||
|
||||
\end_inset
|
||||
|
||||
.
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Standard
|
||||
The statistic
|
||||
\emph on
|
||||
agreement
|
||||
\emph default
|
||||
,
|
||||
\begin_inset Formula $a$
|
||||
\end_inset
|
||||
|
||||
, is defined as the proportion of models under the meta-classifier that
|
||||
correctly predict a sample's class when the ensemble correctly classifies.
|
||||
It could also be considered the confidence of the meta-classifier, for
|
||||
one horizontal model
|
||||
\begin_inset Formula $a_{m=1}=1$
|
||||
\end_inset
|
||||
|
||||
.
|
||||
As error rates are presented, this is inverted by
|
||||
\begin_inset Formula $1-a$
|
||||
\end_inset
|
||||
|
||||
to
|
||||
\emph on
|
||||
disagreement
|
||||
\emph default
|
||||
,
|
||||
\begin_inset Formula $d$
|
||||
\end_inset
|
||||
|
||||
, the proportion of incorrect models when correctly group classifying.
|
||||
\end_layout
|
||||
|
||||
\begin_layout Subsection
|
||||
Results
|
||||
\end_layout
|
||||
|
||||
\begin_layout Standard
|
||||
For comparison, the average individual accuracy for both test and training
|
||||
data are presented.
|
||||
\end_layout
|
||||
|
||||
\begin_layout Standard
|
||||
\begin_inset Float figure
|
||||
wide false
|
||||
sideways false
|
||||
status open
|
||||
|
||||
\begin_layout Plain Layout
|
||||
\noindent
|
||||
\align center
|
||||
\begin_inset Graphics
|
||||
filename ../graphs/exp2-test8-error-rate-curves.png
|
||||
lyxscale 50
|
||||
width 50col%
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Plain Layout
|
||||
\begin_inset Caption Standard
|
||||
|
||||
\begin_layout Plain Layout
|
||||
Ensemble classifier performance results for
|
||||
\begin_inset Formula $\eta=0.03$
|
||||
\end_inset
|
||||
|
||||
,
|
||||
\begin_inset Formula $p=0.01$
|
||||
\end_inset
|
||||
|
||||
, nodes = 1 - 400, epochs = 5 - 100
|
||||
\begin_inset CommandInset label
|
||||
LatexCommand label
|
||||
name "fig:exp2-test8"
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Standard
|
||||
An experiment with a fixed epoch value throughout the ensemble is presented
|
||||
in figure
|
||||
\begin_inset CommandInset ref
|
||||
LatexCommand ref
|
||||
reference "fig:exp2-test10"
|
||||
plural "false"
|
||||
caps "false"
|
||||
noprefix "false"
|
||||
|
||||
\end_inset
|
||||
|
||||
.
|
||||
Nodes between 1 and 400 were selected for the classifiers with a learning
|
||||
rate,
|
||||
\begin_inset Formula $\eta=0.15$
|
||||
\end_inset
|
||||
|
||||
and momentum,
|
||||
\begin_inset Formula $p=0.01$
|
||||
\end_inset
|
||||
|
||||
.
|
||||
The ensemble accuracy can be seen to be fairly constant throughout the
|
||||
number of horizontal models with 3 models being the least accurate with
|
||||
a higher standard deviation.
|
||||
3 horizontal models also shows a significant spike in disagreement and
|
||||
individual error rates which gradually decreases as the number of models
|
||||
increases.
|
||||
\end_layout
|
||||
|
||||
\begin_layout Standard
|
||||
\begin_inset Float figure
|
||||
wide false
|
||||
sideways false
|
||||
status open
|
||||
|
||||
\begin_layout Plain Layout
|
||||
\noindent
|
||||
\align center
|
||||
\begin_inset Graphics
|
||||
filename ../graphs/exp2-test10-error-rate-curves.png
|
||||
lyxscale 50
|
||||
width 50col%
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Plain Layout
|
||||
\begin_inset Caption Standard
|
||||
|
||||
\begin_layout Plain Layout
|
||||
Ensemble classifier performance results for
|
||||
\begin_inset Formula $\eta=0.15$
|
||||
\end_inset
|
||||
|
||||
,
|
||||
\begin_inset Formula $p=0.01$
|
||||
\end_inset
|
||||
|
||||
, nodes =
|
||||
\begin_inset Formula $1-400$
|
||||
\end_inset
|
||||
|
||||
, epochs = 20
|
||||
\begin_inset CommandInset label
|
||||
LatexCommand label
|
||||
name "fig:exp2-test10"
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Plain Layout
|
||||
|
||||
\end_layout
|
||||
|
||||
\end_inset
|
||||
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Subsection
|
||||
Discussion
|
||||
\end_layout
|
||||
|
||||
\begin_layout Standard
|
||||
From the data of figure
|
||||
\begin_inset CommandInset ref
|
||||
LatexCommand ref
|
||||
reference "fig:exp2-test10"
|
||||
plural "false"
|
||||
caps "false"
|
||||
noprefix "false"
|
||||
|
||||
\end_inset
|
||||
|
||||
, 3 horizontal models was shown to be the worst performing configuration
|
||||
with lower ensemble accuracy and higher disagreement.
|
||||
This is likely due to larger proportion that a single model constitutes.
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Section
|
||||
Optimiser Comparisons (Exp 3)
|
||||
Optimiser Comparisons
|
||||
\begin_inset CommandInset label
|
||||
LatexCommand label
|
||||
name "sec:exp3"
|
||||
@ -486,6 +945,20 @@ name "sec:exp3"
|
||||
|
||||
\end_layout
|
||||
|
||||
\begin_layout Standard
|
||||
Throughout the previous experiments the stochastic gradient descent optimiser
|
||||
was used to change the networks weights but there are many different optimisati
|
||||
on algorithms.
|
||||
This section will present investigations into two other optimisation algorithms
|
||||
and discuss the differences between them using the horizontal ensemble
|
||||
classification of the previous section.
|
||||
\end_layout
|
||||
|
||||
\begin_layout Standard
|
||||
Prior to these investigations, however, stochastic gradient descent and
|
||||
the two other subject algorithms will be described.
|
||||
\end_layout
|
||||
|
||||
\begin_layout Subsection
|
||||
Optimisers
|
||||
\end_layout
|
||||
@ -510,10 +983,6 @@ Results
|
||||
Discussion
|
||||
\end_layout
|
||||
|
||||
\begin_layout Section
|
||||
Overlapping 2D Gaussians (Exp 4)
|
||||
\end_layout
|
||||
|
||||
\begin_layout Section
|
||||
Conclusions
|
||||
\end_layout
|
||||
|
||||